RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
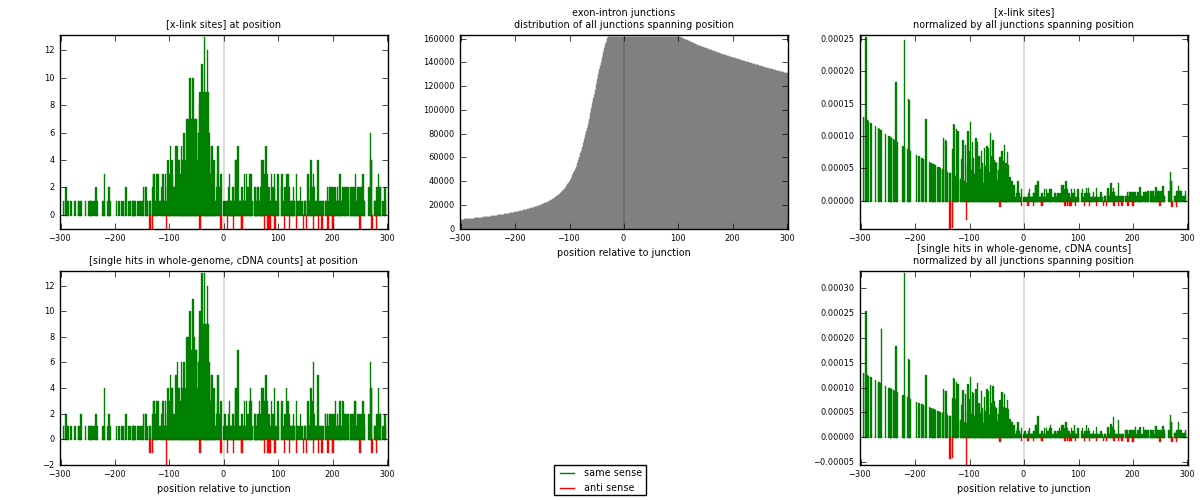 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 866 | 5.16% | 19.92% | 96.76% | 742 | 1.12468 |
| anti | 29 | 0.17% | 0.67% | 3.24% | 28 | 0.0376623 |
| both | 895 | 5.33% | 20.59% | 100.00% | 770 | 1.16234 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 904 | 5.02% | 18.32% | 96.79% | 742 | 1.17403 |
| anti | 30 | 0.17% | 0.61% | 3.21% | 28 | 0.038961 |
| both | 934 | 5.18% | 18.93% | 100.00% | 770 | 1.21299 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
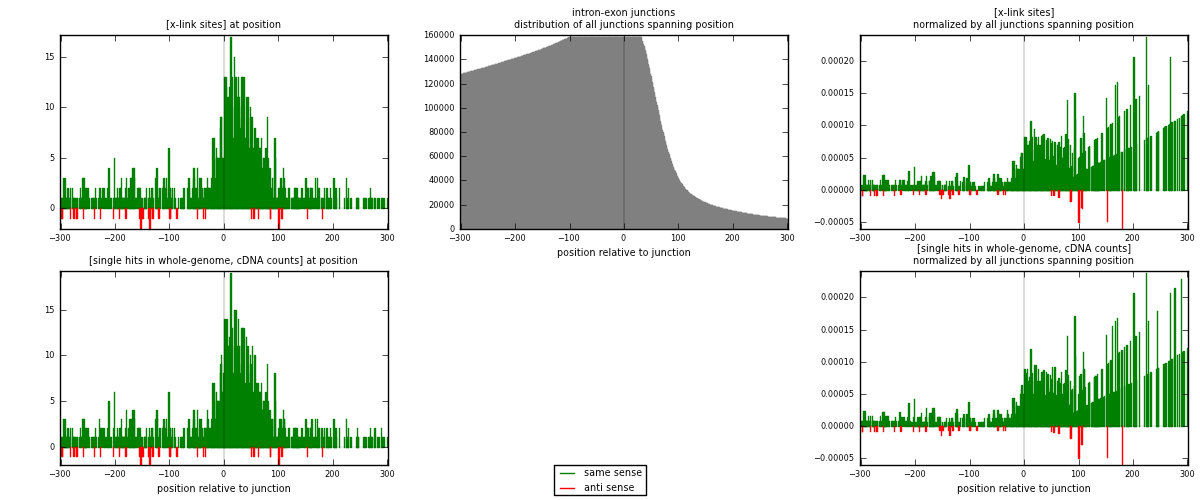 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1193 | 7.11% | 27.44% | 96.76% | 1000 | 1.15154 |
| anti | 40 | 0.24% | 0.92% | 3.24% | 36 | 0.03861 |
| both | 1233 | 7.35% | 28.36% | 100.00% | 1036 | 1.19015 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1255 | 6.97% | 25.44% | 96.91% | 1000 | 1.21139 |
| anti | 40 | 0.22% | 0.81% | 3.09% | 36 | 0.03861 |
| both | 1295 | 7.19% | 26.25% | 100.00% | 1036 | 1.25 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
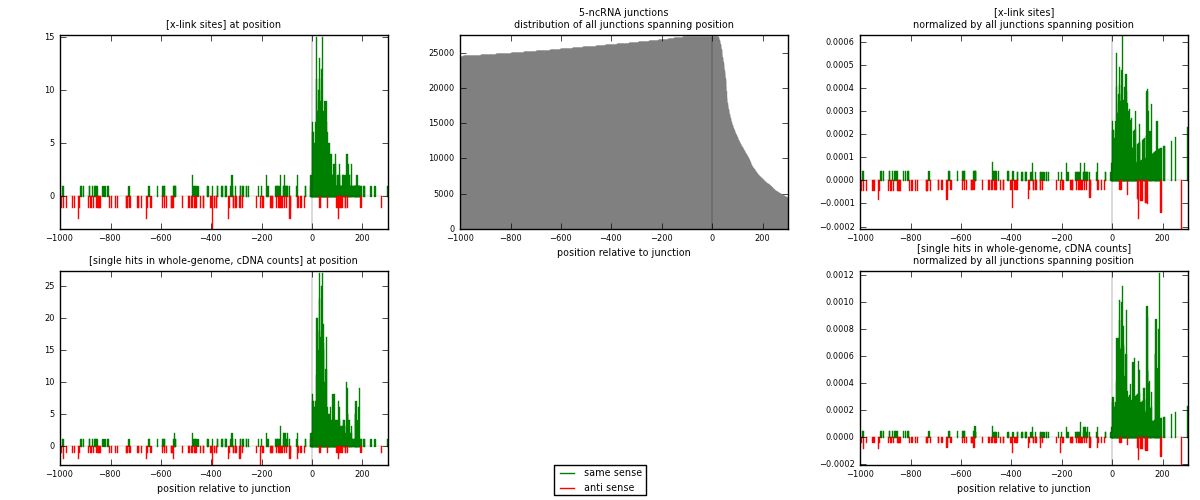 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 639 | 3.81% | 14.70% | 85.77% | 329 | 1.53237 |
| anti | 106 | 0.63% | 2.44% | 14.23% | 88 | 0.254197 |
| both | 745 | 4.44% | 17.14% | 100.00% | 417 | 1.78657 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 948 | 5.26% | 19.21% | 89.43% | 329 | 2.27338 |
| anti | 112 | 0.62% | 2.27% | 10.57% | 88 | 0.268585 |
| both | 1060 | 5.88% | 21.48% | 100.00% | 417 | 2.54197 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
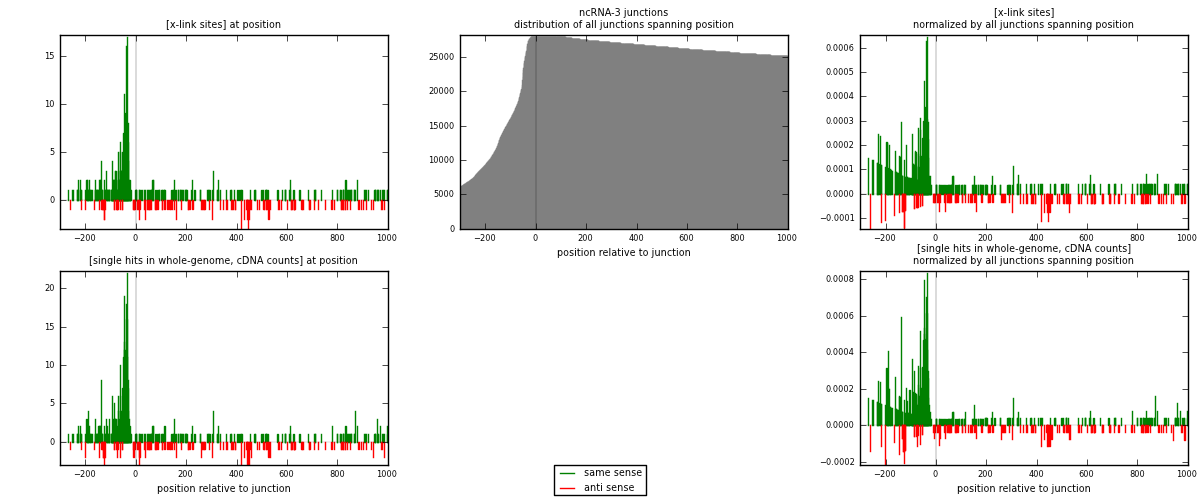 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 447 | 2.66% | 10.28% | 75.63% | 243 | 1.34234 |
| anti | 144 | 0.86% | 3.31% | 24.37% | 93 | 0.432432 |
| both | 591 | 3.52% | 13.60% | 100.00% | 333 | 1.77477 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 566 | 3.14% | 11.47% | 77.64% | 243 | 1.6997 |
| anti | 163 | 0.90% | 3.30% | 22.36% | 93 | 0.489489 |
| both | 729 | 4.05% | 14.78% | 100.00% | 333 | 2.18919 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
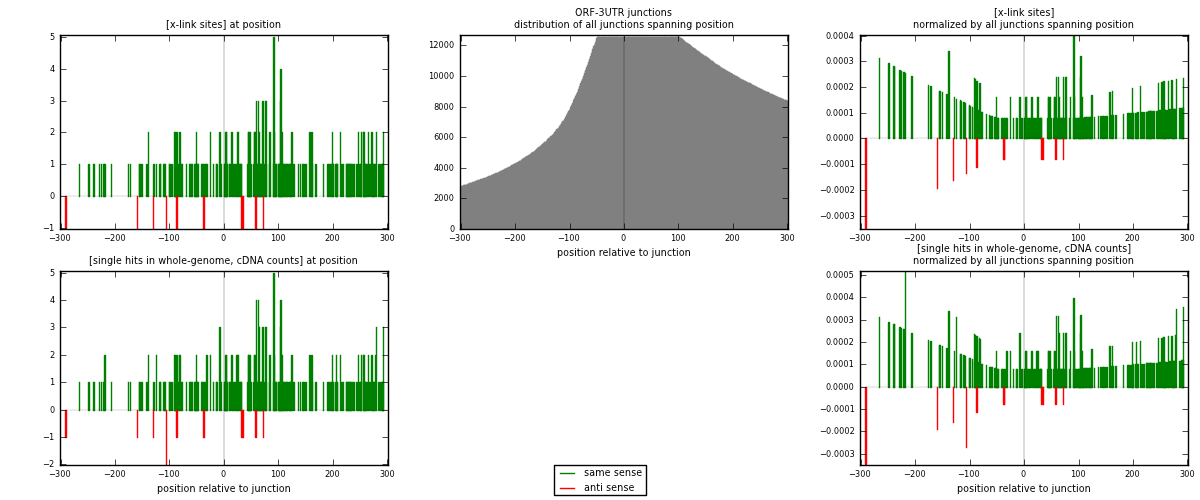 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 259 | 1.54% | 5.96% | 95.93% | 203 | 1.24519 |
| anti | 11 | 0.07% | 0.25% | 4.07% | 5 | 0.0528846 |
| both | 270 | 1.61% | 6.21% | 100.00% | 208 | 1.29808 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 272 | 1.51% | 5.51% | 95.77% | 203 | 1.30769 |
| anti | 12 | 0.07% | 0.24% | 4.23% | 5 | 0.0576923 |
| both | 284 | 1.58% | 5.76% | 100.00% | 208 | 1.36538 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
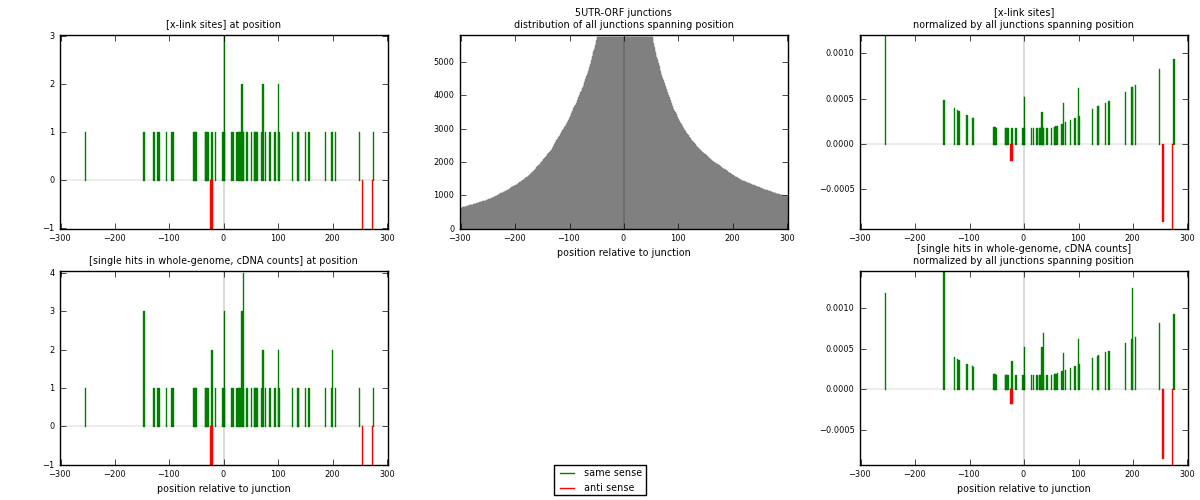 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 59 | 0.35% | 1.36% | 92.19% | 45 | 1.25532 |
| anti | 5 | 0.03% | 0.12% | 7.81% | 2 | 0.106383 |
| both | 64 | 0.38% | 1.47% | 100.00% | 47 | 1.3617 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 67 | 0.37% | 1.36% | 93.06% | 45 | 1.42553 |
| anti | 5 | 0.03% | 0.10% | 6.94% | 2 | 0.106383 |
| both | 72 | 0.40% | 1.46% | 100.00% | 47 | 1.53191 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
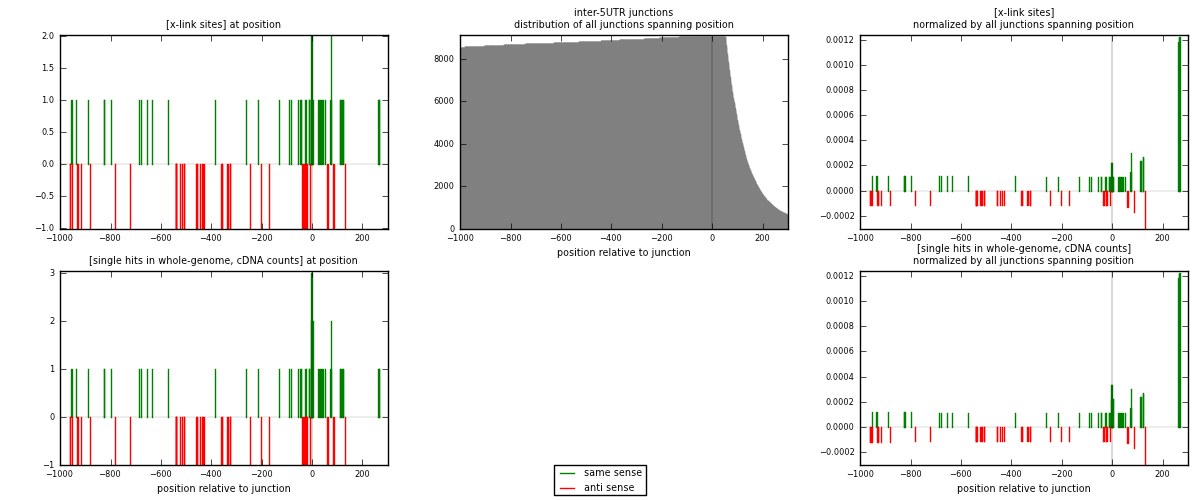 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 48 | 0.29% | 1.10% | 55.17% | 32 | 0.8 |
| anti | 39 | 0.23% | 0.90% | 44.83% | 28 | 0.65 |
| both | 87 | 0.52% | 2.00% | 100.00% | 60 | 1.45 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 53 | 0.29% | 1.07% | 57.61% | 32 | 0.883333 |
| anti | 39 | 0.22% | 0.79% | 42.39% | 28 | 0.65 |
| both | 92 | 0.51% | 1.86% | 100.00% | 60 | 1.53333 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
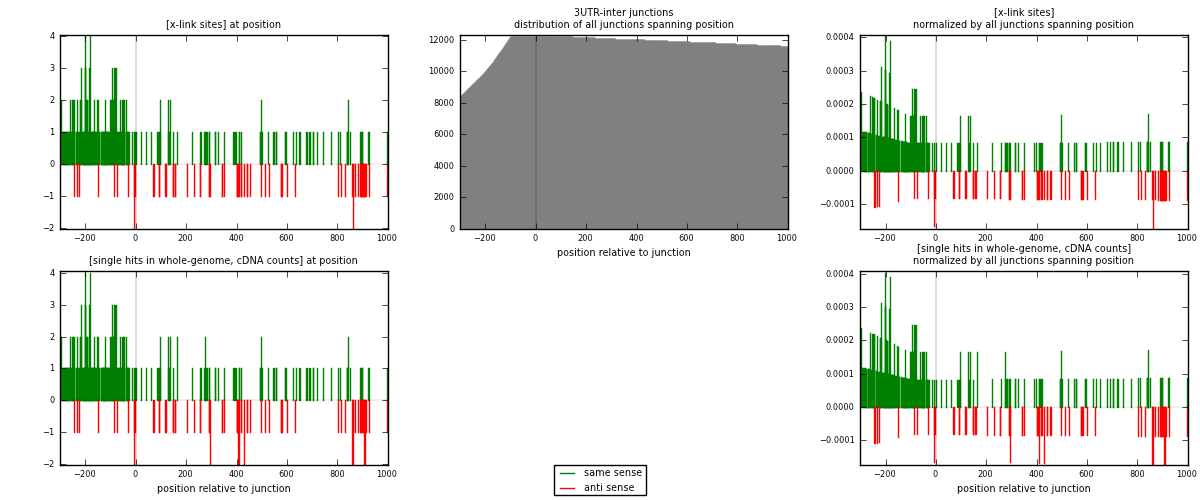 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 252 | 1.50% | 5.80% | 78.26% | 197 | 1.03279 |
| anti | 70 | 0.42% | 1.61% | 21.74% | 47 | 0.286885 |
| both | 322 | 1.92% | 7.41% | 100.00% | 244 | 1.31967 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 254 | 1.41% | 5.15% | 76.97% | 197 | 1.04098 |
| anti | 76 | 0.42% | 1.54% | 23.03% | 47 | 0.311475 |
| both | 330 | 1.83% | 6.69% | 100.00% | 244 | 1.35246 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
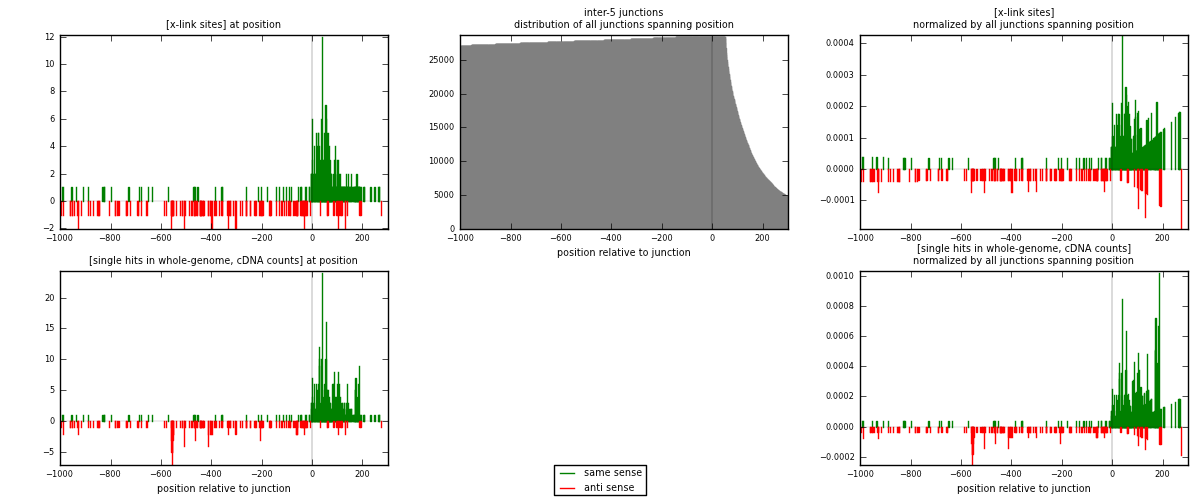 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 348 | 2.07% | 8.01% | 68.10% | 196 | 1.2 |
| anti | 163 | 0.97% | 3.75% | 31.90% | 94 | 0.562069 |
| both | 511 | 3.05% | 11.76% | 100.00% | 290 | 1.76207 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 520 | 2.89% | 10.54% | 72.32% | 196 | 1.7931 |
| anti | 199 | 1.10% | 4.03% | 27.68% | 94 | 0.686207 |
| both | 719 | 3.99% | 14.57% | 100.00% | 290 | 2.47931 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
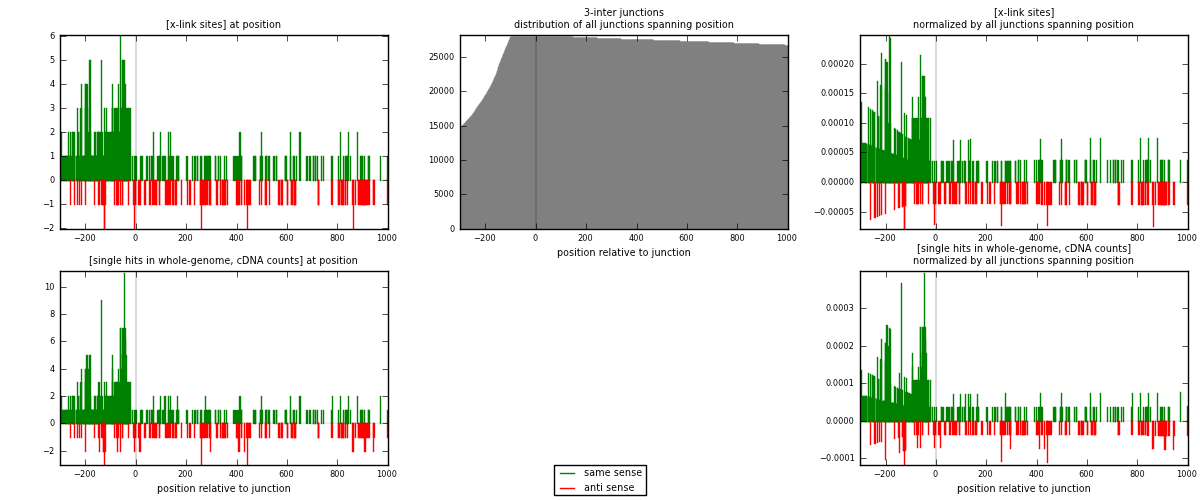 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 447 | 2.66% | 10.28% | 73.28% | 297 | 1.1175 |
| anti | 163 | 0.97% | 3.75% | 26.72% | 104 | 0.4075 |
| both | 610 | 3.64% | 14.03% | 100.00% | 400 | 1.525 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 504 | 2.80% | 10.21% | 73.47% | 297 | 1.26 |
| anti | 182 | 1.01% | 3.69% | 26.53% | 104 | 0.455 |
| both | 686 | 3.81% | 13.90% | 100.00% | 400 | 1.715 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
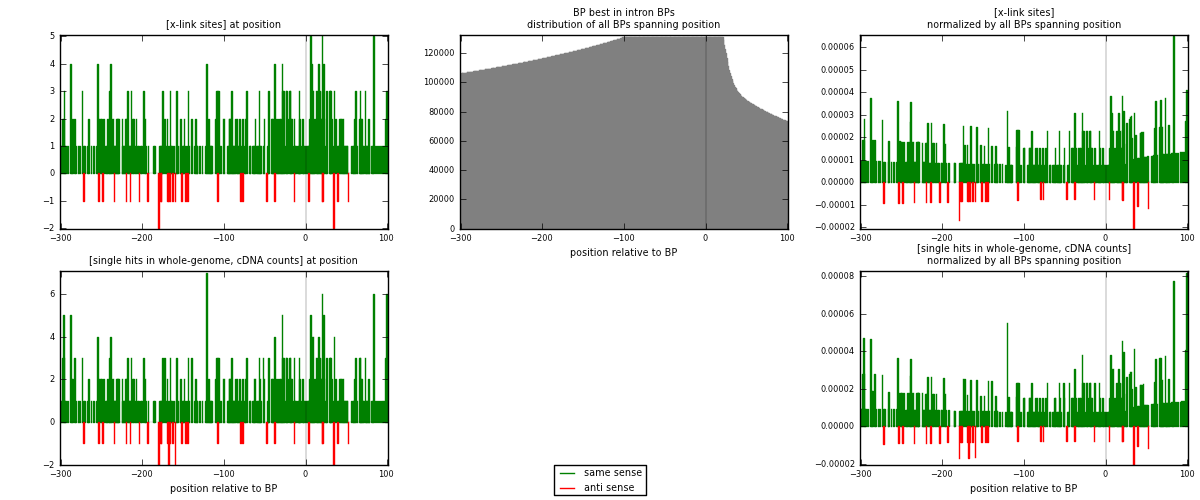 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 456 | 2.72% | 10.49% | 93.25% | 342 | 1.22911 |
| anti | 33 | 0.20% | 0.76% | 6.75% | 29 | 0.0889488 |
| both | 489 | 2.91% | 11.25% | 100.00% | 371 | 1.31806 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 485 | 2.69% | 9.83% | 93.27% | 342 | 1.30728 |
| anti | 35 | 0.19% | 0.71% | 6.73% | 29 | 0.0943396 |
| both | 520 | 2.89% | 10.54% | 100.00% | 371 | 1.40162 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.