RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
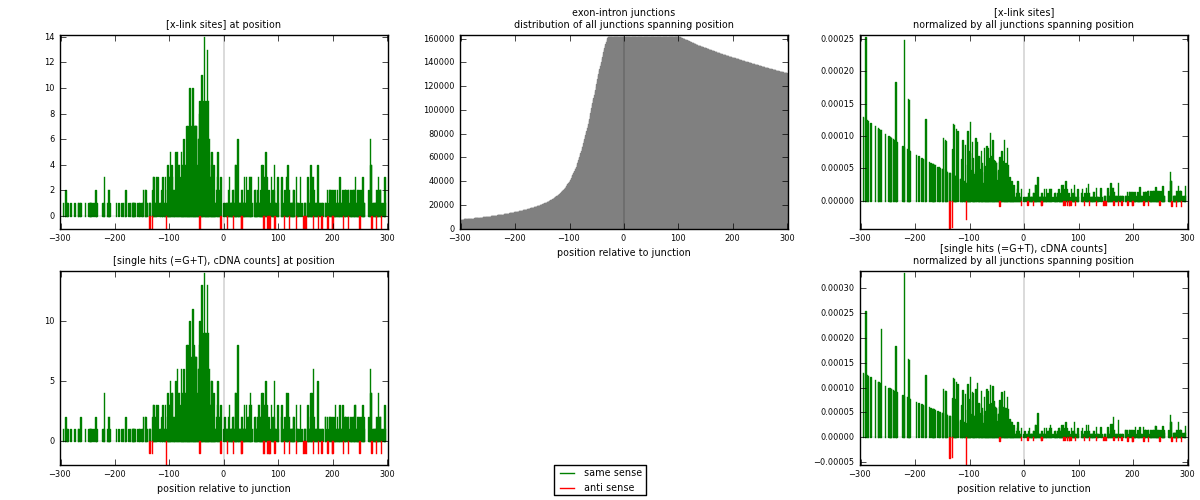 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 889 | 4.67% | 18.88% | 96.32% | 762 | 1.11824 |
| anti | 34 | 0.18% | 0.72% | 3.68% | 33 | 0.0427673 |
| both | 923 | 4.85% | 19.60% | 100.00% | 795 | 1.16101 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 927 | 4.46% | 16.74% | 96.36% | 762 | 1.16604 |
| anti | 35 | 0.17% | 0.63% | 3.64% | 33 | 0.0440252 |
| both | 962 | 4.63% | 17.37% | 100.00% | 795 | 1.21006 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
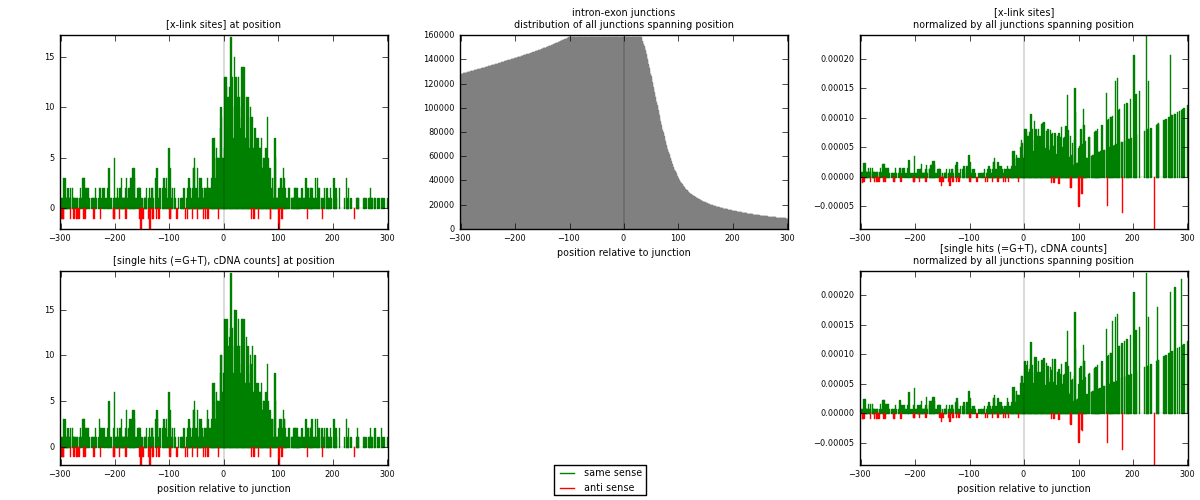 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1212 | 6.37% | 25.74% | 95.73% | 1017 | 1.1359 |
| anti | 54 | 0.28% | 1.15% | 4.27% | 50 | 0.0506092 |
| both | 1266 | 6.66% | 26.89% | 100.00% | 1067 | 1.1865 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1274 | 6.13% | 23.00% | 95.93% | 1017 | 1.194 |
| anti | 54 | 0.26% | 0.98% | 4.07% | 50 | 0.0506092 |
| both | 1328 | 6.39% | 23.98% | 100.00% | 1067 | 1.24461 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
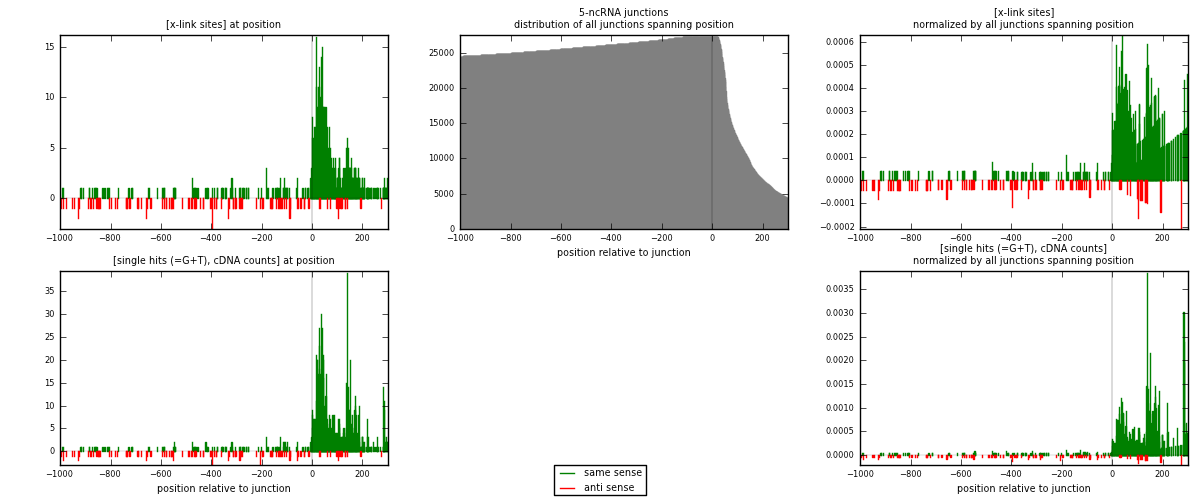 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 822 | 4.32% | 17.46% | 87.91% | 353 | 1.83893 |
| anti | 113 | 0.59% | 2.40% | 12.09% | 94 | 0.252796 |
| both | 935 | 4.92% | 19.86% | 100.00% | 447 | 2.09172 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1359 | 6.54% | 24.54% | 91.95% | 353 | 3.04027 |
| anti | 119 | 0.57% | 2.15% | 8.05% | 94 | 0.266219 |
| both | 1478 | 7.11% | 26.69% | 100.00% | 447 | 3.30649 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
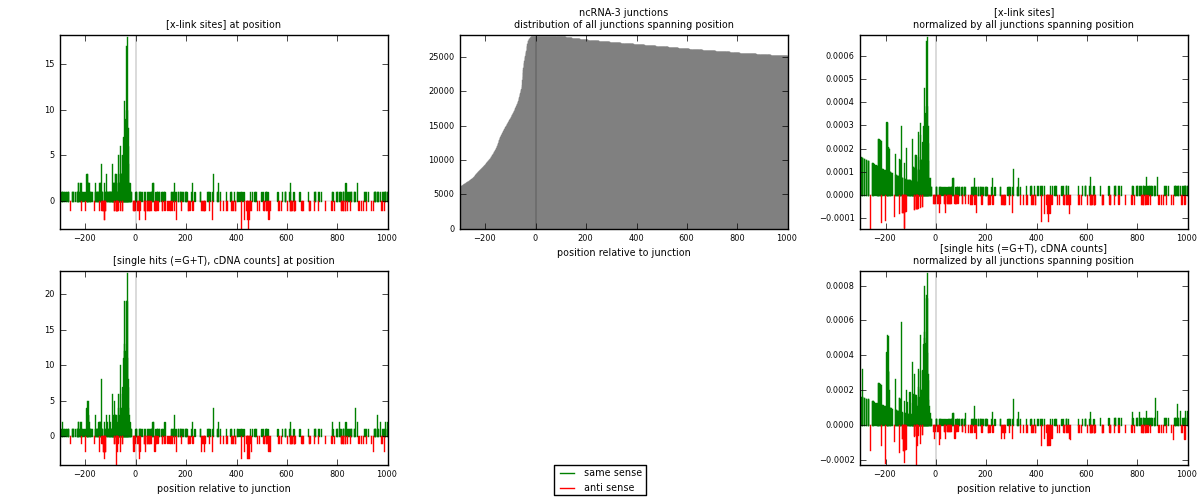 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 506 | 2.66% | 10.75% | 76.55% | 258 | 1.42938 |
| anti | 155 | 0.81% | 3.29% | 23.45% | 99 | 0.437853 |
| both | 661 | 3.47% | 14.04% | 100.00% | 354 | 1.86723 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 636 | 3.06% | 11.48% | 77.94% | 258 | 1.79661 |
| anti | 180 | 0.87% | 3.25% | 22.06% | 99 | 0.508475 |
| both | 816 | 3.93% | 14.73% | 100.00% | 354 | 2.30508 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
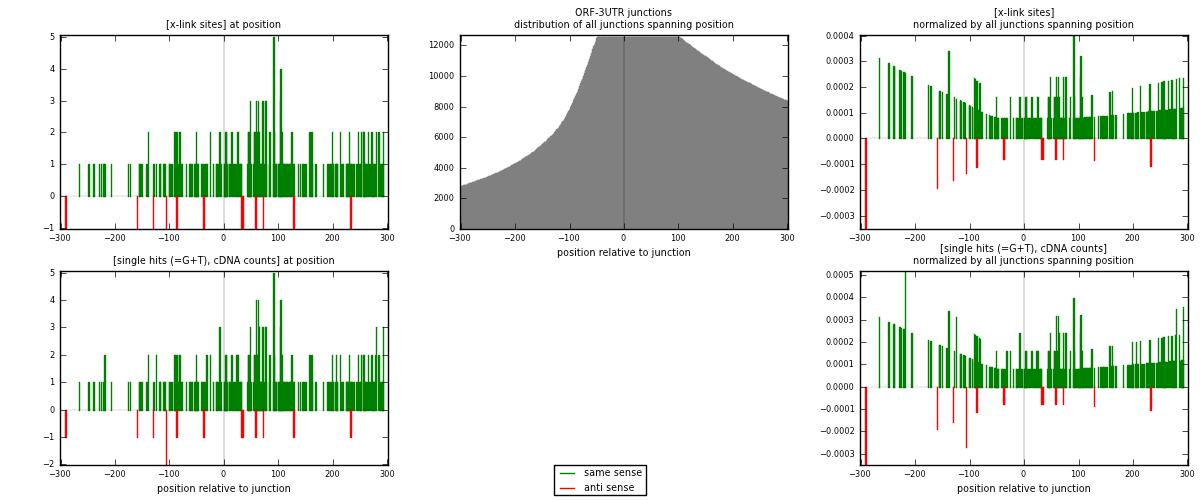 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 264 | 1.39% | 5.61% | 95.31% | 206 | 1.23944 |
| anti | 13 | 0.07% | 0.28% | 4.69% | 7 | 0.0610329 |
| both | 277 | 1.46% | 5.88% | 100.00% | 213 | 1.30047 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 277 | 1.33% | 5.00% | 95.19% | 206 | 1.30047 |
| anti | 14 | 0.07% | 0.25% | 4.81% | 7 | 0.0657277 |
| both | 291 | 1.40% | 5.25% | 100.00% | 213 | 1.3662 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
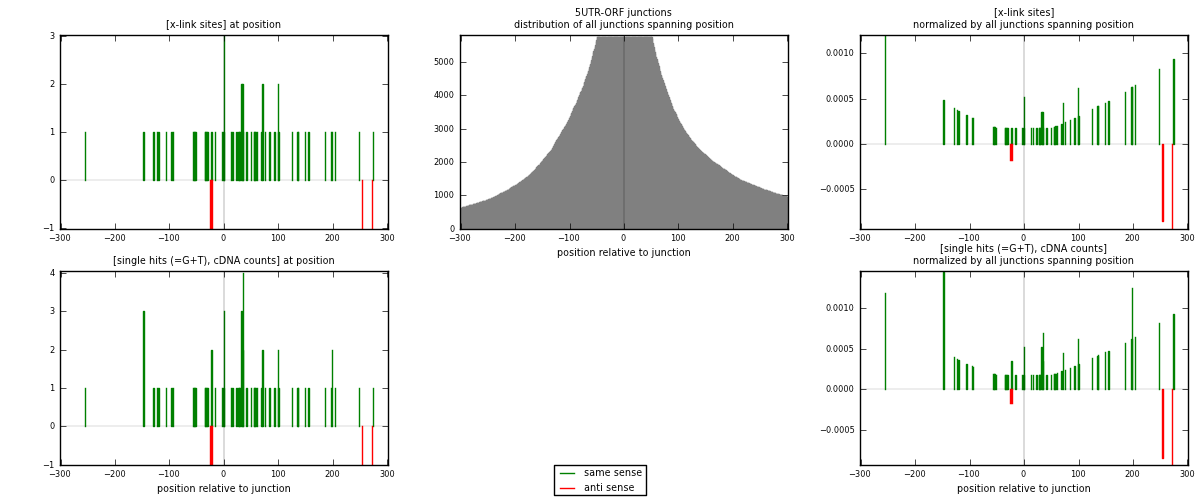 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 60 | 0.32% | 1.27% | 92.31% | 45 | 1.2766 |
| anti | 5 | 0.03% | 0.11% | 7.69% | 2 | 0.106383 |
| both | 65 | 0.34% | 1.38% | 100.00% | 47 | 1.38298 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 68 | 0.33% | 1.23% | 93.15% | 45 | 1.44681 |
| anti | 5 | 0.02% | 0.09% | 6.85% | 2 | 0.106383 |
| both | 73 | 0.35% | 1.32% | 100.00% | 47 | 1.55319 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
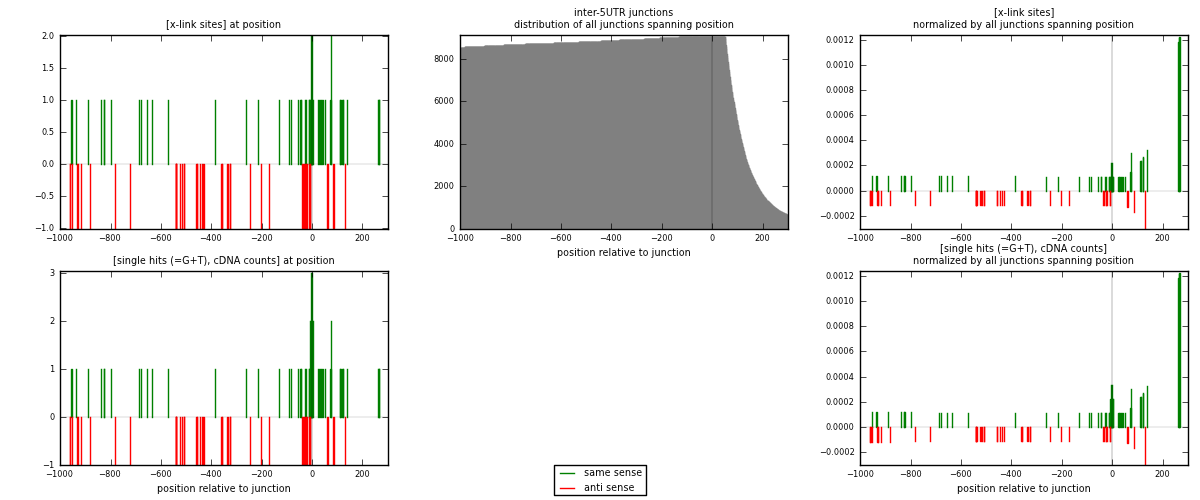 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 51 | 0.27% | 1.08% | 56.04% | 34 | 0.809524 |
| anti | 40 | 0.21% | 0.85% | 43.96% | 29 | 0.634921 |
| both | 91 | 0.48% | 1.93% | 100.00% | 63 | 1.44444 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 57 | 0.27% | 1.03% | 58.76% | 34 | 0.904762 |
| anti | 40 | 0.19% | 0.72% | 41.24% | 29 | 0.634921 |
| both | 97 | 0.47% | 1.75% | 100.00% | 63 | 1.53968 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 254 | 1.34% | 5.40% | 77.20% | 198 | 1.016 |
| anti | 75 | 0.39% | 1.59% | 22.80% | 52 | 0.3 |
| both | 329 | 1.73% | 6.99% | 100.00% | 250 | 1.316 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 256 | 1.23% | 4.62% | 75.96% | 198 | 1.024 |
| anti | 81 | 0.39% | 1.46% | 24.04% | 52 | 0.324 |
| both | 337 | 1.62% | 6.09% | 100.00% | 250 | 1.348 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
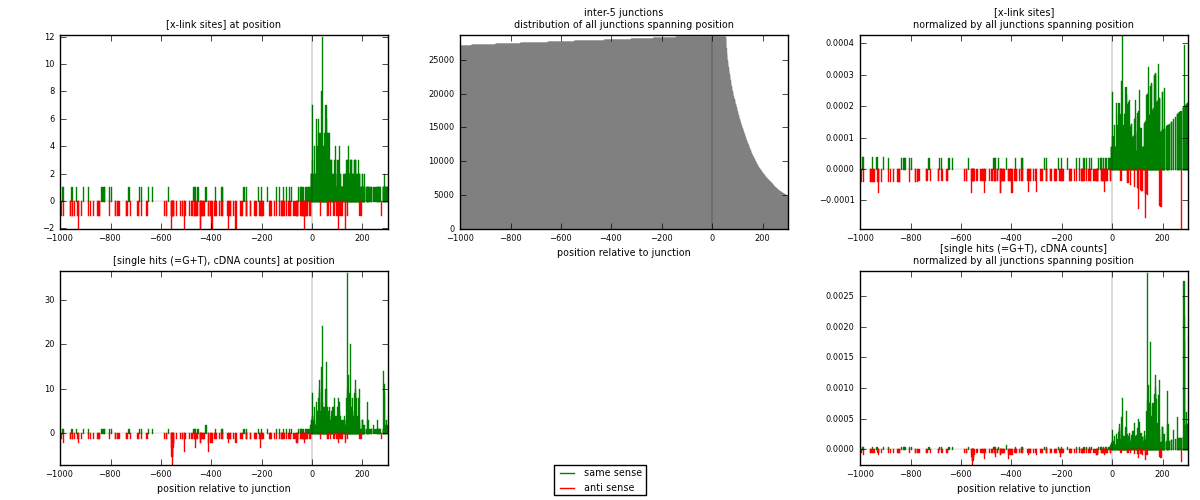 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 520 | 2.73% | 11.05% | 74.93% | 210 | 1.67203 |
| anti | 174 | 0.91% | 3.70% | 25.07% | 101 | 0.559486 |
| both | 694 | 3.65% | 14.74% | 100.00% | 311 | 2.23151 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 920 | 4.43% | 16.61% | 81.42% | 210 | 2.9582 |
| anti | 210 | 1.01% | 3.79% | 18.58% | 101 | 0.675241 |
| both | 1130 | 5.44% | 20.40% | 100.00% | 311 | 3.63344 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
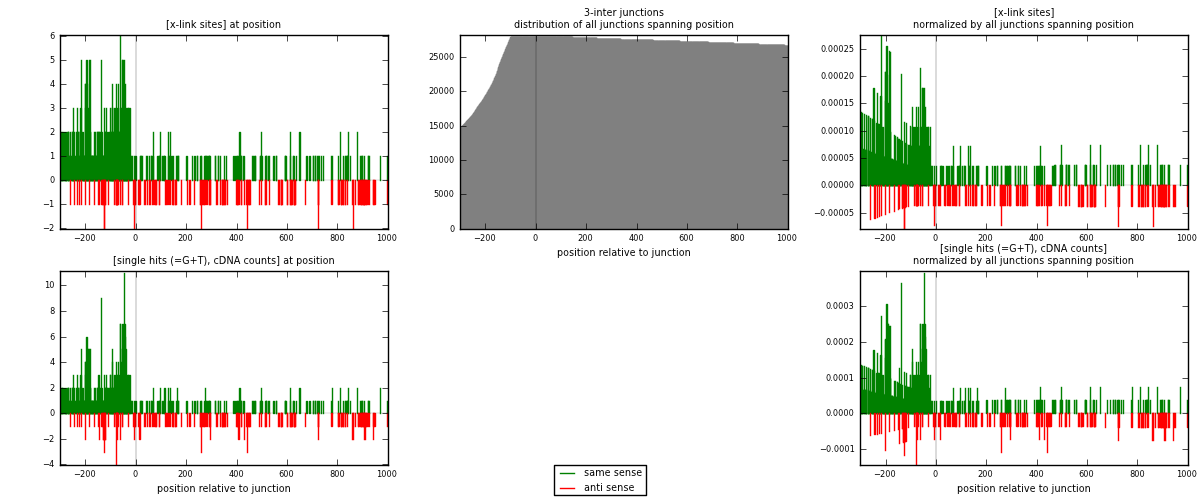 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 494 | 2.60% | 10.49% | 73.73% | 305 | 1.1875 |
| anti | 176 | 0.93% | 3.74% | 26.27% | 112 | 0.423077 |
| both | 670 | 3.52% | 14.23% | 100.00% | 416 | 1.61058 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 559 | 2.69% | 10.09% | 73.75% | 305 | 1.34375 |
| anti | 199 | 0.96% | 3.59% | 26.25% | 112 | 0.478365 |
| both | 758 | 3.65% | 13.69% | 100.00% | 416 | 1.82212 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
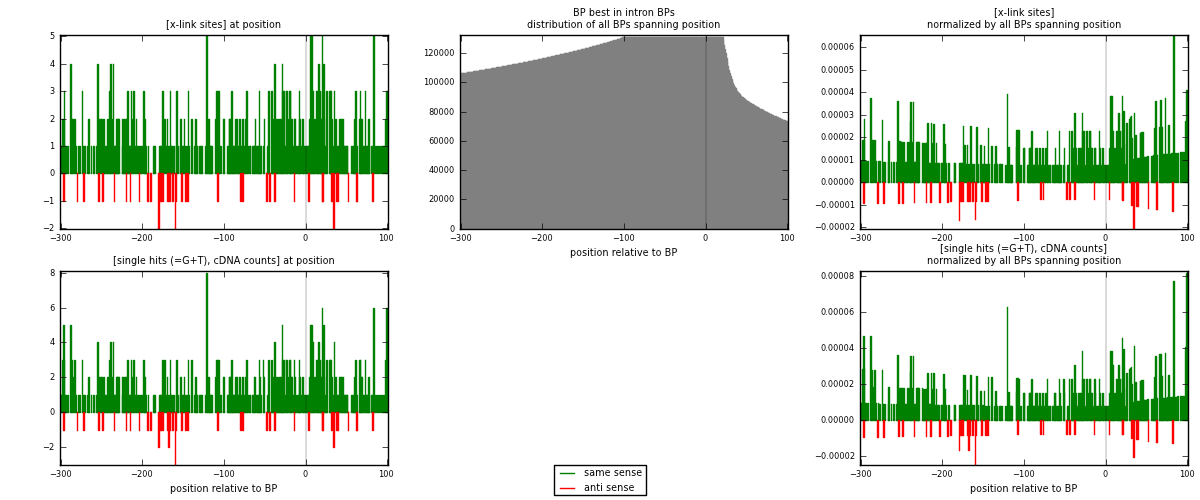 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 470 | 2.47% | 9.98% | 91.26% | 355 | 1.18987 |
| anti | 45 | 0.24% | 0.96% | 8.74% | 40 | 0.113924 |
| both | 515 | 2.71% | 10.94% | 100.00% | 395 | 1.3038 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 499 | 2.40% | 9.01% | 91.39% | 355 | 1.26329 |
| anti | 47 | 0.23% | 0.85% | 8.61% | 40 | 0.118987 |
| both | 546 | 2.63% | 9.86% | 100.00% | 395 | 1.38228 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.