RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
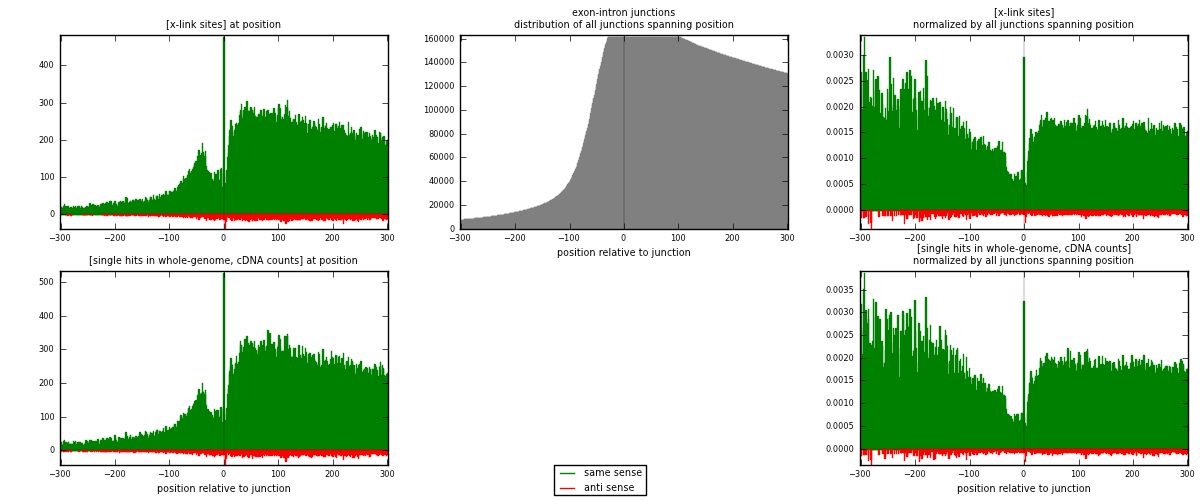 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 86281 | 3.56% | 27.31% | 95.59% | 36725 | 2.22868 |
| anti | 3984 | 0.16% | 1.26% | 4.41% | 2379 | 0.102909 |
| both | 90265 | 3.72% | 28.57% | 100.00% | 38714 | 2.33159 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 97537 | 3.52% | 25.27% | 95.76% | 36725 | 2.51942 |
| anti | 4319 | 0.16% | 1.12% | 4.24% | 2379 | 0.111562 |
| both | 101856 | 3.68% | 26.39% | 100.00% | 38714 | 2.63099 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
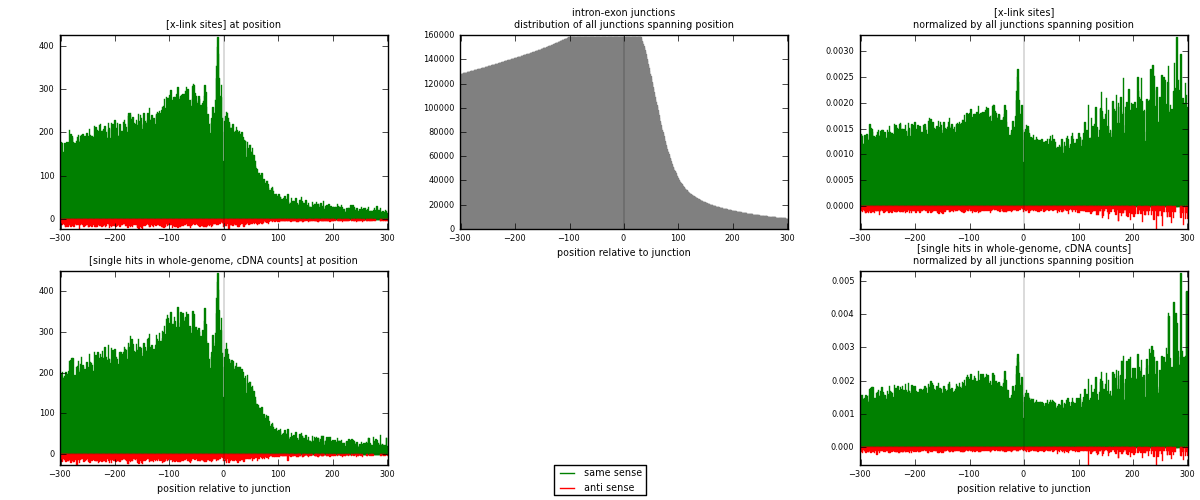 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 87480 | 3.61% | 27.69% | 95.23% | 38035 | 2.17196 |
| anti | 4382 | 0.18% | 1.39% | 4.77% | 2738 | 0.108797 |
| both | 91862 | 3.79% | 29.07% | 100.00% | 40277 | 2.28076 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 98282 | 3.55% | 25.46% | 95.35% | 38035 | 2.44015 |
| anti | 4789 | 0.17% | 1.24% | 4.65% | 2738 | 0.118902 |
| both | 103071 | 3.72% | 26.70% | 100.00% | 40277 | 2.55905 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
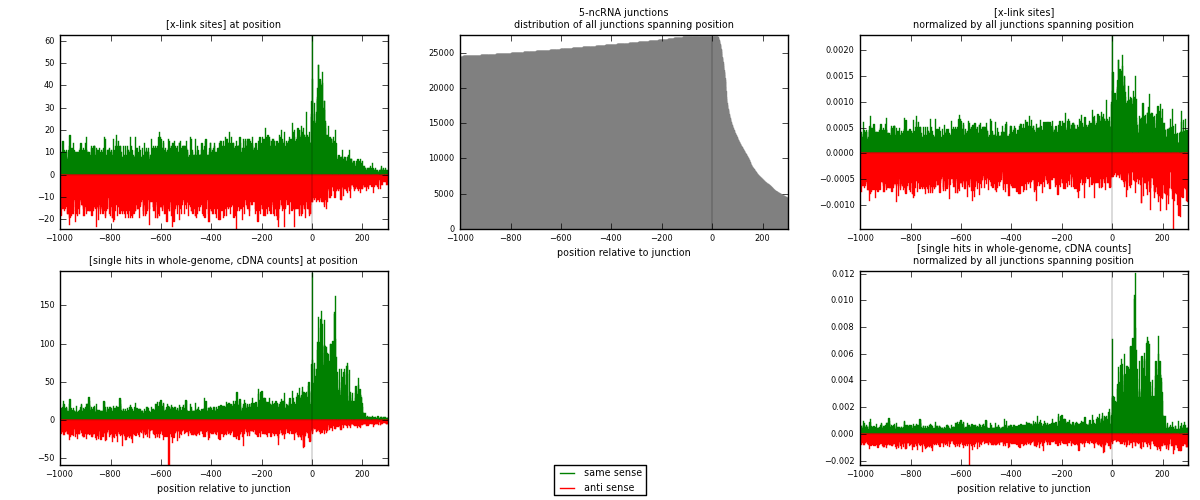 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13053 | 0.54% | 4.13% | 48.51% | 3101 | 1.99161 |
| anti | 13853 | 0.57% | 4.38% | 51.49% | 3696 | 2.11367 |
| both | 26906 | 1.11% | 8.52% | 100.00% | 6554 | 4.10528 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 24046 | 0.87% | 6.23% | 60.43% | 3101 | 3.6689 |
| anti | 15744 | 0.57% | 4.08% | 39.57% | 3696 | 2.4022 |
| both | 39790 | 1.44% | 10.31% | 100.00% | 6554 | 6.0711 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
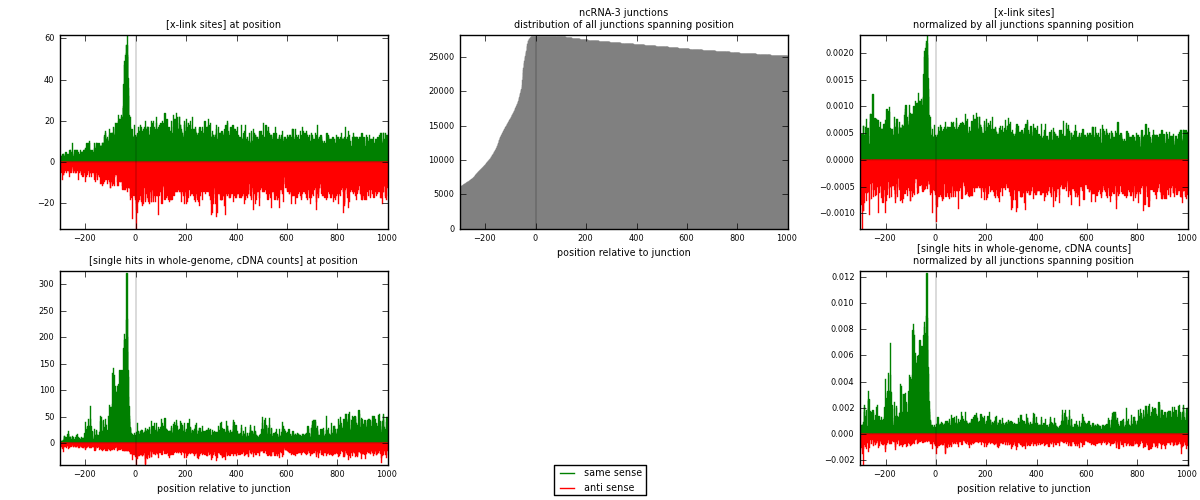 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 14119 | 0.58% | 4.47% | 49.68% | 3407 | 2.08091 |
| anti | 14303 | 0.59% | 4.53% | 50.32% | 3635 | 2.10803 |
| both | 28422 | 1.17% | 9.00% | 100.00% | 6785 | 4.18895 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 32491 | 1.17% | 8.42% | 66.23% | 3407 | 4.78865 |
| anti | 16569 | 0.60% | 4.29% | 33.77% | 3635 | 2.442 |
| both | 49060 | 1.77% | 12.71% | 100.00% | 6785 | 7.23066 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
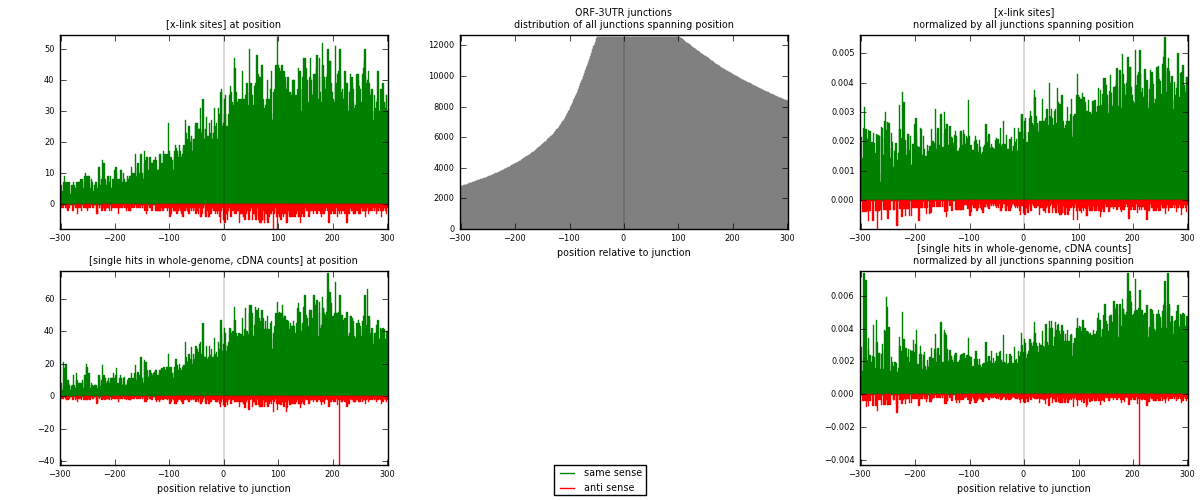
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 14119 | 0.58% | 4.47% | 93.71% | 4190 | 3.10581 |
| anti | 948 | 0.04% | 0.30% | 6.29% | 484 | 0.208535 |
| both | 15067 | 0.62% | 4.77% | 100.00% | 4546 | 3.31434 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 16881 | 0.61% | 4.37% | 94.17% | 4190 | 3.71337 |
| anti | 1045 | 0.04% | 0.27% | 5.83% | 484 | 0.229872 |
| both | 17926 | 0.65% | 4.64% | 100.00% | 4546 | 3.94325 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
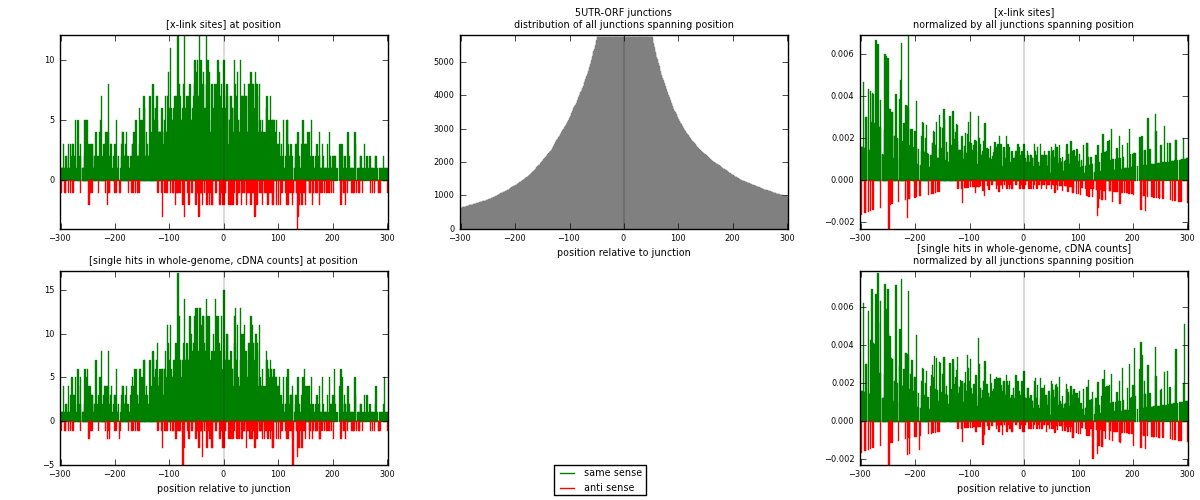
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1873 | 0.08% | 0.59% | 88.68% | 768 | 2.20612 |
| anti | 239 | 0.01% | 0.08% | 11.32% | 90 | 0.281508 |
| both | 2112 | 0.09% | 0.67% | 100.00% | 849 | 2.48763 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2240 | 0.08% | 0.58% | 89.10% | 768 | 2.6384 |
| anti | 274 | 0.01% | 0.07% | 10.90% | 90 | 0.322733 |
| both | 2514 | 0.09% | 0.65% | 100.00% | 849 | 2.96113 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
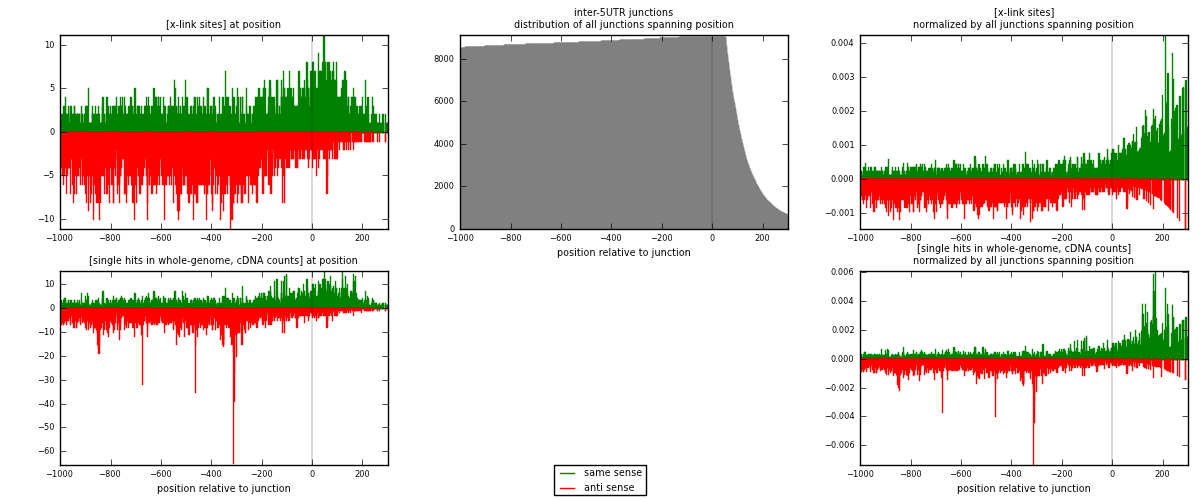
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2616 | 0.11% | 0.83% | 39.56% | 874 | 1.29249 |
| anti | 3996 | 0.16% | 1.26% | 60.44% | 1279 | 1.97431 |
| both | 6612 | 0.27% | 2.09% | 100.00% | 2024 | 3.2668 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3221 | 0.12% | 0.83% | 39.77% | 874 | 1.5914 |
| anti | 4878 | 0.18% | 1.26% | 60.23% | 1279 | 2.41008 |
| both | 8099 | 0.29% | 2.10% | 100.00% | 2024 | 4.00148 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
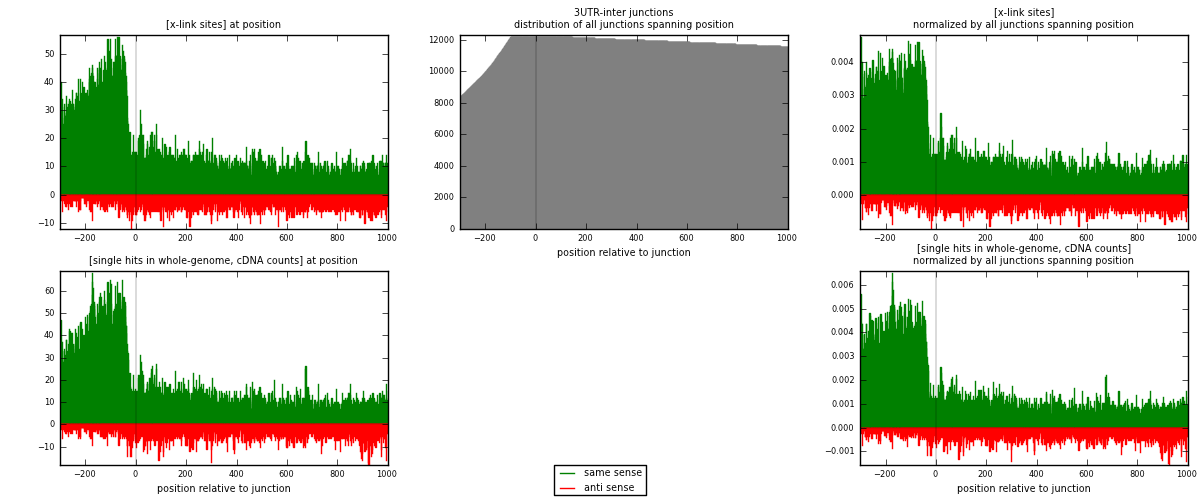
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 19335 | 0.80% | 6.12% | 80.24% | 4999 | 3.23652 |
| anti | 4760 | 0.20% | 1.51% | 19.76% | 1512 | 0.796786 |
| both | 24095 | 0.99% | 7.63% | 100.00% | 5974 | 4.03331 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 21708 | 0.78% | 5.62% | 79.67% | 4999 | 3.63375 |
| anti | 5540 | 0.20% | 1.44% | 20.33% | 1512 | 0.927352 |
| both | 27248 | 0.98% | 7.06% | 100.00% | 5974 | 4.5611 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
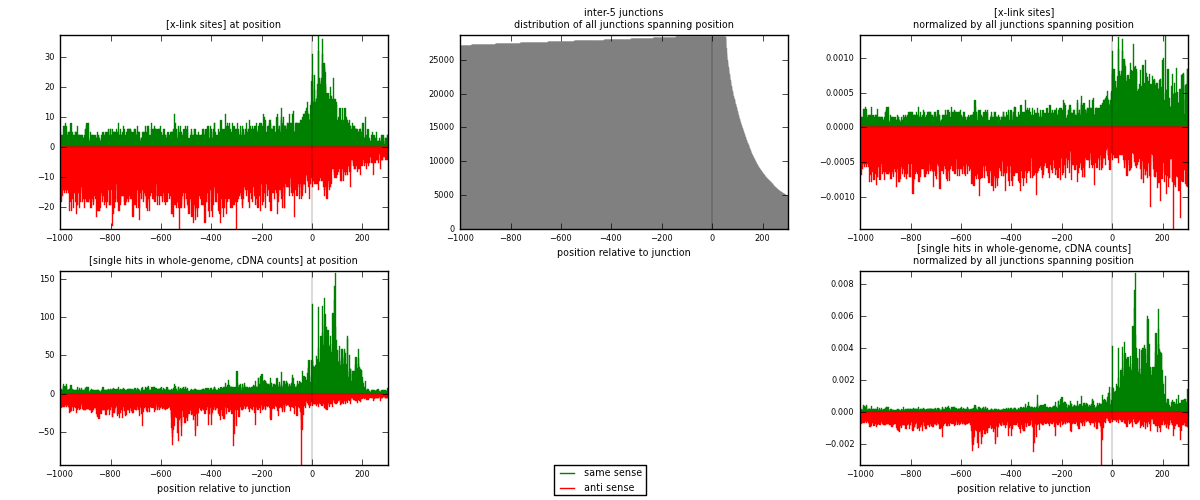
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6516 | 0.27% | 2.06% | 29.86% | 2150 | 1.06037 |
| anti | 15303 | 0.63% | 4.84% | 70.14% | 4301 | 2.49032 |
| both | 21819 | 0.90% | 6.91% | 100.00% | 6145 | 3.55069 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 13979 | 0.51% | 3.62% | 42.71% | 2150 | 2.27486 |
| anti | 18752 | 0.68% | 4.86% | 57.29% | 4301 | 3.05159 |
| both | 32731 | 1.18% | 8.48% | 100.00% | 6145 | 5.32644 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
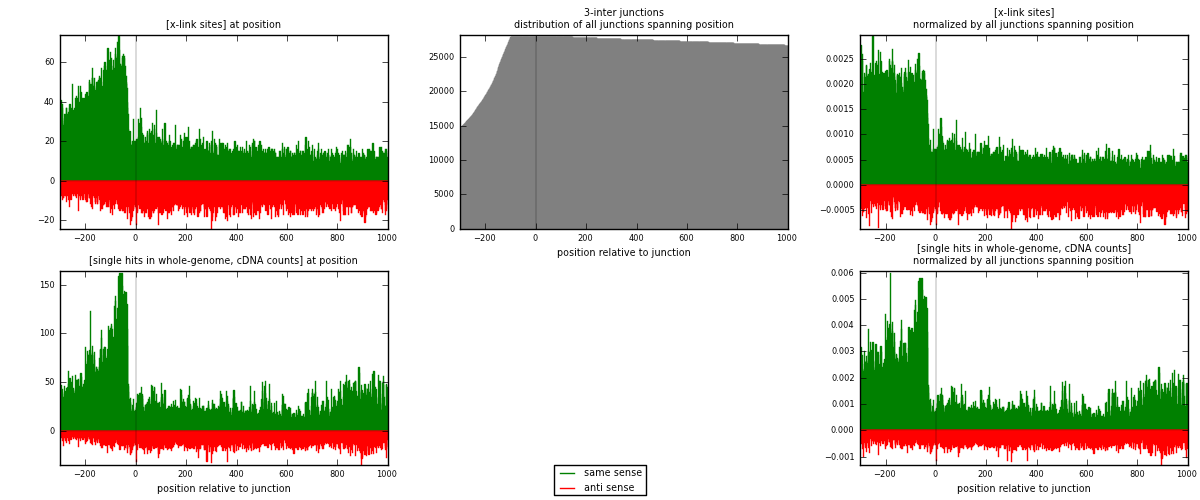
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 25533 | 1.05% | 8.08% | 65.08% | 6587 | 2.63444 |
| anti | 13702 | 0.56% | 4.34% | 34.92% | 3840 | 1.41374 |
| both | 39235 | 1.62% | 12.42% | 100.00% | 9692 | 4.04818 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 41552 | 1.50% | 10.76% | 72.68% | 6587 | 4.28725 |
| anti | 15618 | 0.56% | 4.05% | 27.32% | 3840 | 1.61143 |
| both | 57170 | 2.07% | 14.81% | 100.00% | 9692 | 5.89868 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
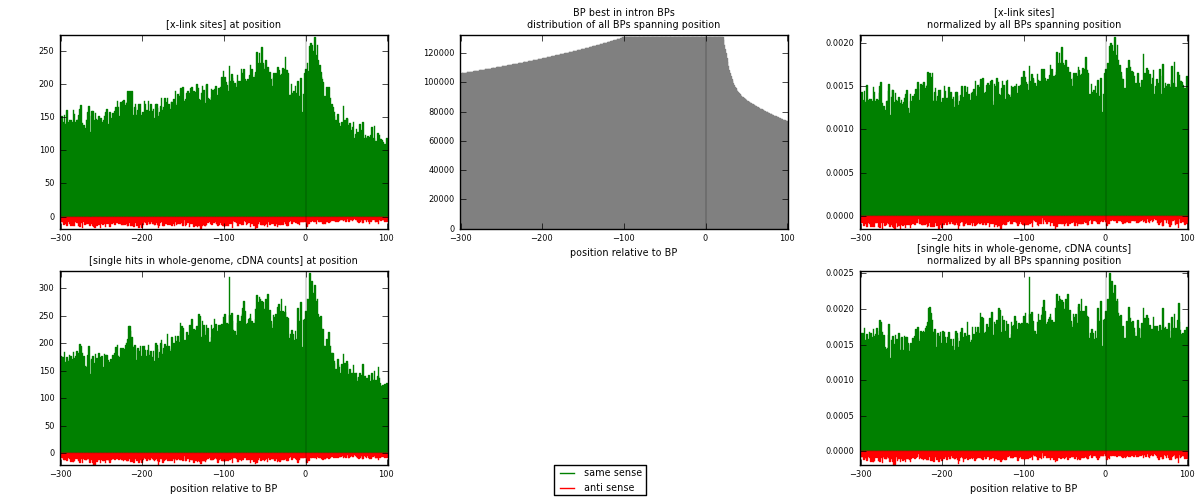
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 68740 | 2.83% | 21.76% | 95.55% | 28839 | 2.24692 |
| anti | 3204 | 0.13% | 1.01% | 4.45% | 2090 | 0.10473 |
| both | 71944 | 2.97% | 22.77% | 100.00% | 30593 | 2.35165 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 79808 | 2.88% | 20.67% | 95.90% | 28839 | 2.6087 |
| anti | 3412 | 0.12% | 0.88% | 4.10% | 2090 | 0.111529 |
| both | 83220 | 3.01% | 21.56% | 100.00% | 30593 | 2.72023 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.