RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
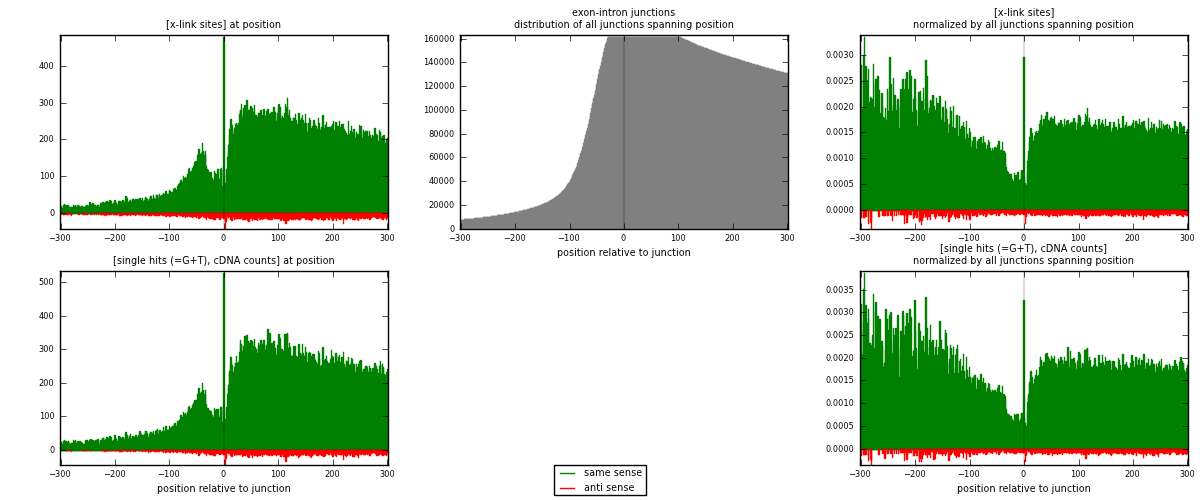 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 87129 | 3.52% | 27.09% | 95.43% | 37045 | 2.22614 |
| anti | 4173 | 0.17% | 1.30% | 4.57% | 2512 | 0.10662 |
| both | 91302 | 3.68% | 28.39% | 100.00% | 39139 | 2.33276 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 98546 | 3.47% | 24.95% | 95.61% | 37045 | 2.51785 |
| anti | 4523 | 0.16% | 1.15% | 4.39% | 2512 | 0.115562 |
| both | 103069 | 3.63% | 26.09% | 100.00% | 39139 | 2.63341 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
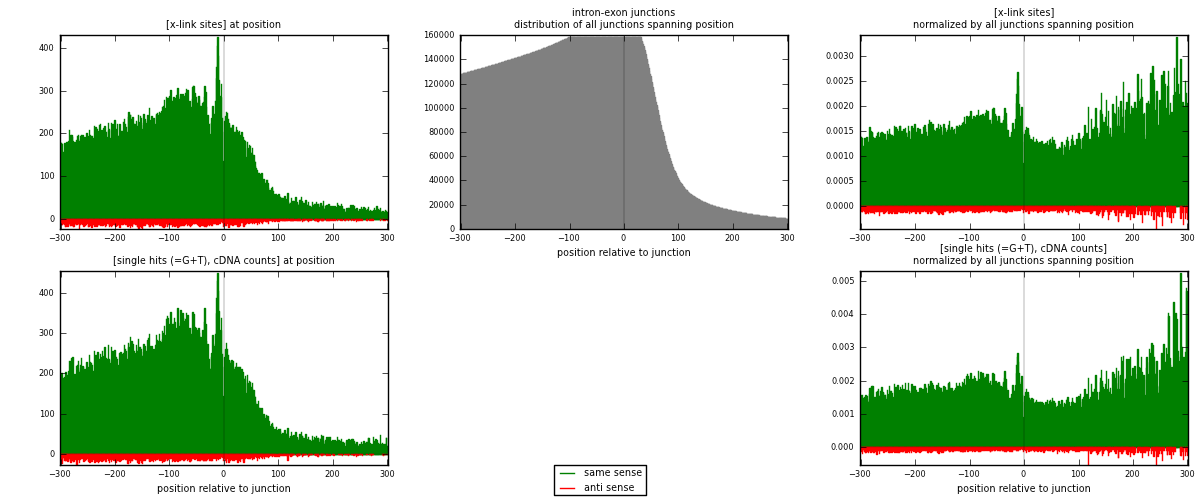 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 88196 | 3.56% | 27.42% | 95.03% | 38278 | 2.16922 |
| anti | 4614 | 0.19% | 1.43% | 4.97% | 2918 | 0.113483 |
| both | 92810 | 3.74% | 28.86% | 100.00% | 40658 | 2.2827 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 99134 | 3.49% | 25.10% | 95.17% | 38278 | 2.43824 |
| anti | 5033 | 0.18% | 1.27% | 4.83% | 2918 | 0.123789 |
| both | 104167 | 3.67% | 26.37% | 100.00% | 40658 | 2.56203 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
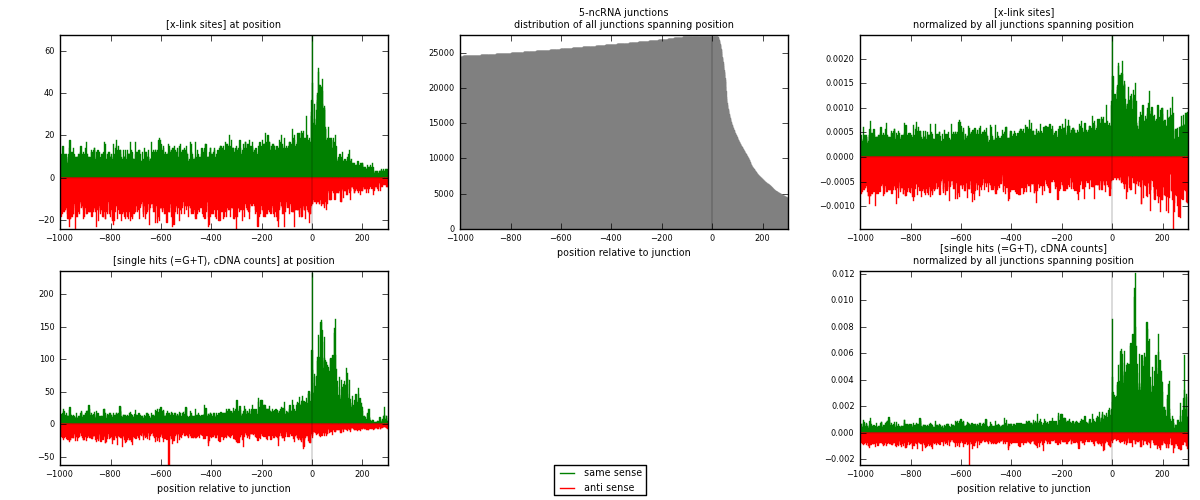 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13733 | 0.55% | 4.27% | 49.06% | 3234 | 2.03001 |
| anti | 14258 | 0.58% | 4.43% | 50.94% | 3807 | 2.10761 |
| both | 27991 | 1.13% | 8.70% | 100.00% | 6765 | 4.13762 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 25751 | 0.91% | 6.52% | 61.23% | 3234 | 3.8065 |
| anti | 16306 | 0.57% | 4.13% | 38.77% | 3807 | 2.41035 |
| both | 42057 | 1.48% | 10.65% | 100.00% | 6765 | 6.21685 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
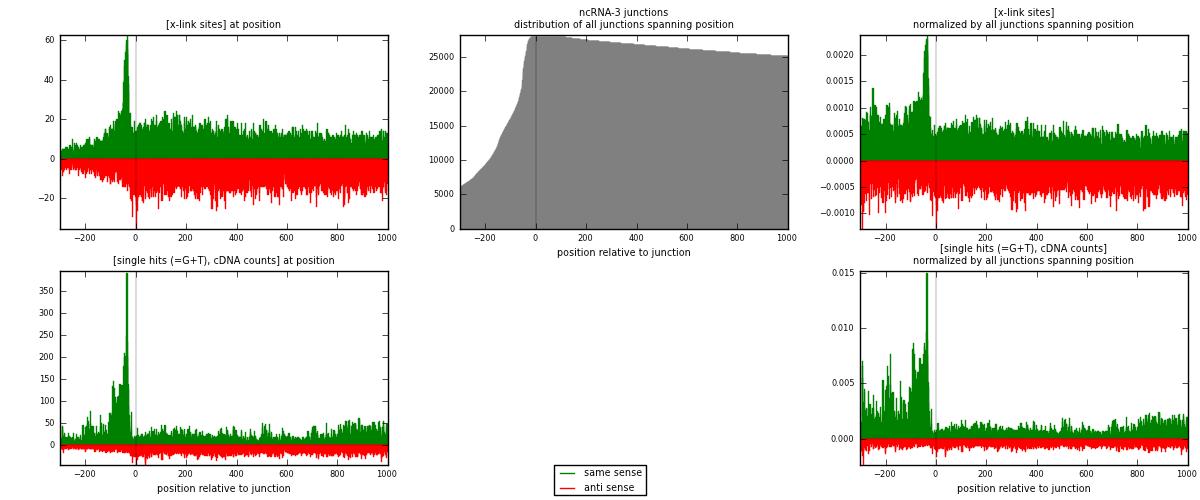 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 14763 | 0.60% | 4.59% | 50.01% | 3519 | 2.11232 |
| anti | 14755 | 0.60% | 4.59% | 49.99% | 3758 | 2.11117 |
| both | 29518 | 1.19% | 9.18% | 100.00% | 6989 | 4.22349 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 34510 | 1.22% | 8.74% | 66.79% | 3519 | 4.93776 |
| anti | 17161 | 0.60% | 4.34% | 33.21% | 3758 | 2.45543 |
| both | 51671 | 1.82% | 13.08% | 100.00% | 6989 | 7.39319 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
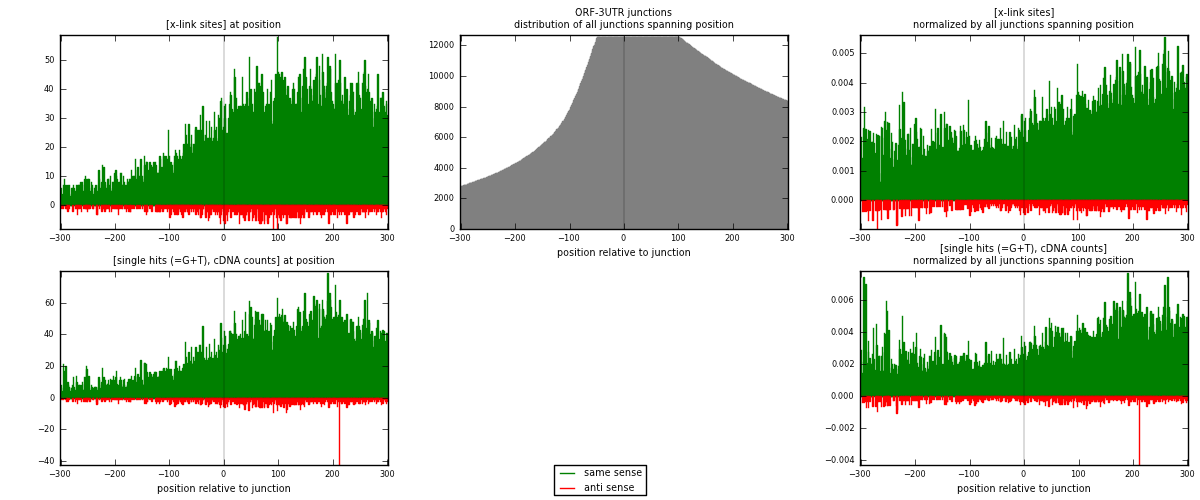 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 14425 | 0.58% | 4.49% | 93.71% | 4239 | 3.13315 |
| anti | 968 | 0.04% | 0.30% | 6.29% | 499 | 0.210252 |
| both | 15393 | 0.62% | 4.79% | 100.00% | 4604 | 3.3434 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 17247 | 0.61% | 4.37% | 94.18% | 4239 | 3.74609 |
| anti | 1065 | 0.04% | 0.27% | 5.82% | 499 | 0.231321 |
| both | 18312 | 0.65% | 4.64% | 100.00% | 4604 | 3.97741 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
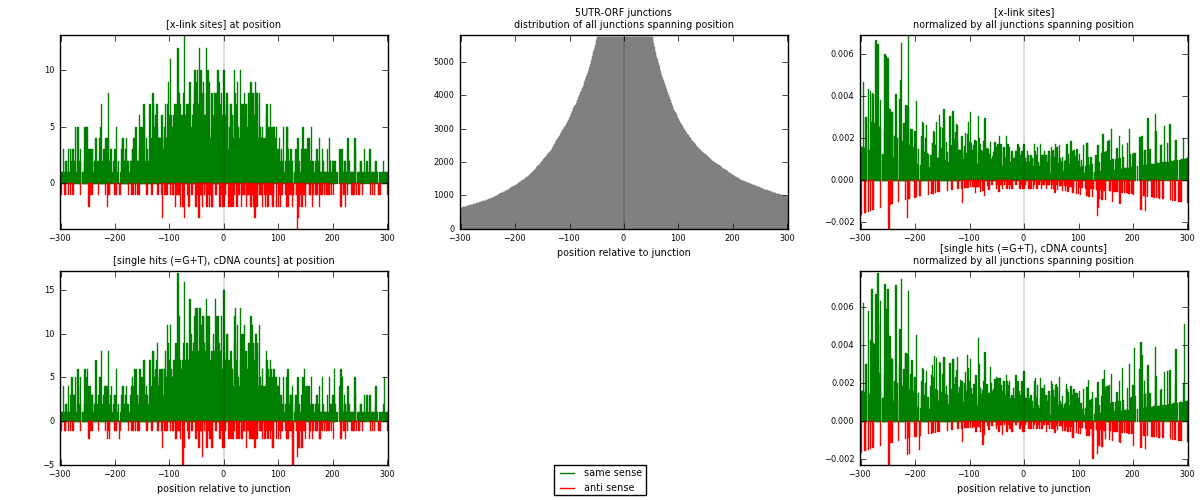 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1883 | 0.08% | 0.59% | 88.49% | 771 | 2.1972 |
| anti | 245 | 0.01% | 0.08% | 11.51% | 95 | 0.285881 |
| both | 2128 | 0.09% | 0.66% | 100.00% | 857 | 2.48308 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2254 | 0.08% | 0.57% | 88.95% | 771 | 2.63011 |
| anti | 280 | 0.01% | 0.07% | 11.05% | 95 | 0.326721 |
| both | 2534 | 0.09% | 0.64% | 100.00% | 857 | 2.95683 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
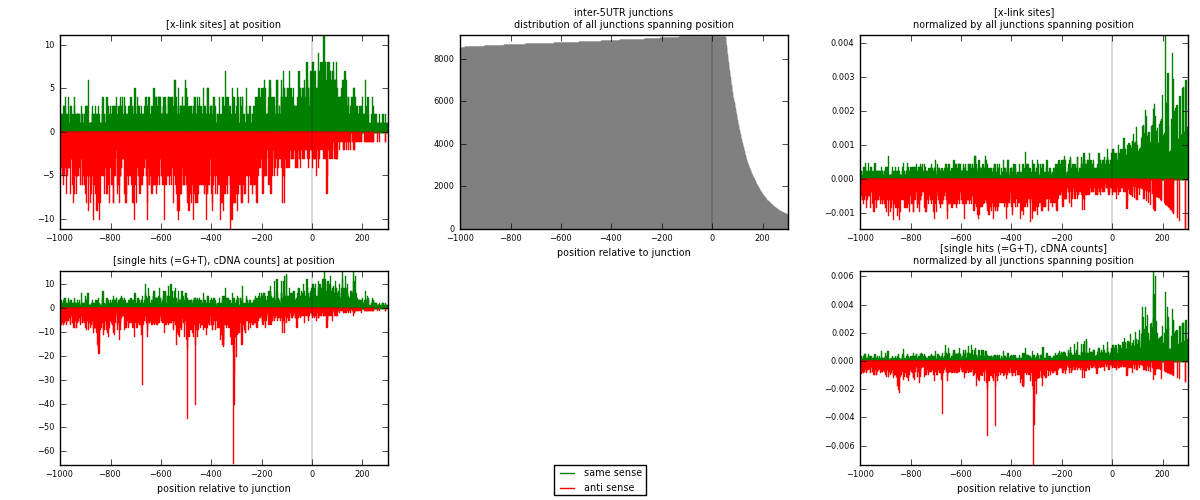 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2717 | 0.11% | 0.84% | 40.13% | 894 | 1.32537 |
| anti | 4054 | 0.16% | 1.26% | 59.87% | 1292 | 1.97756 |
| both | 6771 | 0.27% | 2.11% | 100.00% | 2050 | 3.30293 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3407 | 0.12% | 0.86% | 40.47% | 894 | 1.66195 |
| anti | 5011 | 0.18% | 1.27% | 59.53% | 1292 | 2.44439 |
| both | 8418 | 0.30% | 2.13% | 100.00% | 2050 | 4.10634 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
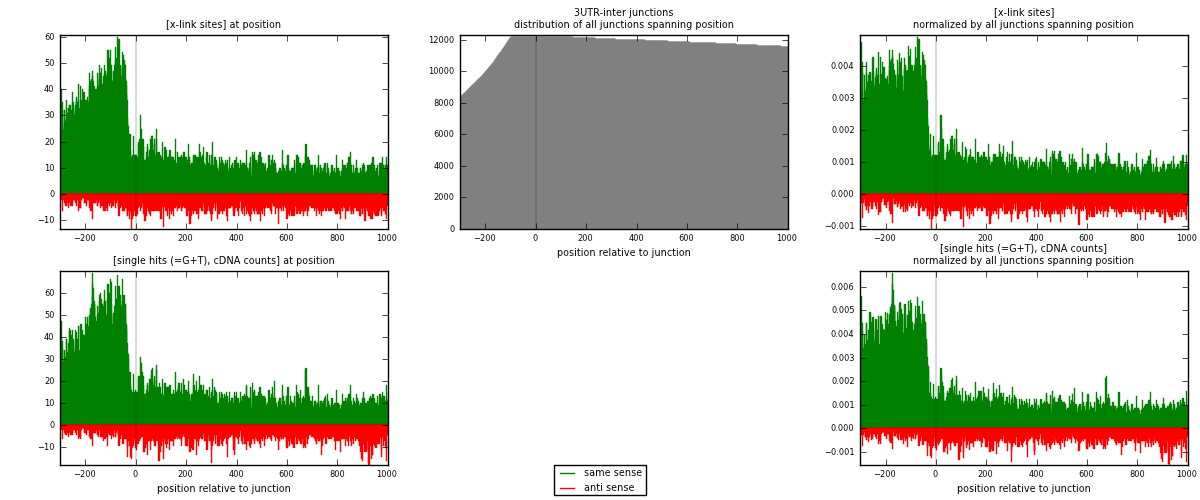 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 19674 | 0.79% | 6.12% | 80.08% | 5057 | 3.2519 |
| anti | 4894 | 0.20% | 1.52% | 19.92% | 1551 | 0.808926 |
| both | 24568 | 0.99% | 7.64% | 100.00% | 6050 | 4.06083 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 22088 | 0.78% | 5.59% | 79.50% | 5057 | 3.65091 |
| anti | 5694 | 0.20% | 1.44% | 20.50% | 1551 | 0.941157 |
| both | 27782 | 0.98% | 7.03% | 100.00% | 6050 | 4.59207 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
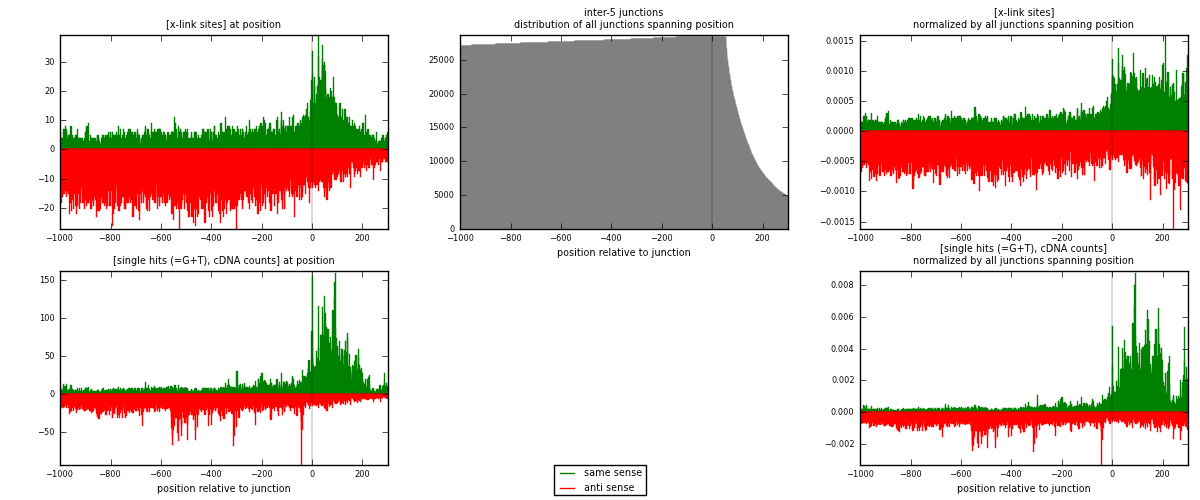 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7099 | 0.29% | 2.21% | 31.32% | 2242 | 1.12736 |
| anti | 15564 | 0.63% | 4.84% | 68.68% | 4388 | 2.47165 |
| both | 22663 | 0.91% | 7.05% | 100.00% | 6297 | 3.59902 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 15511 | 0.55% | 3.93% | 44.79% | 2242 | 2.46324 |
| anti | 19116 | 0.67% | 4.84% | 55.21% | 4388 | 3.03573 |
| both | 34627 | 1.22% | 8.77% | 100.00% | 6297 | 5.49897 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
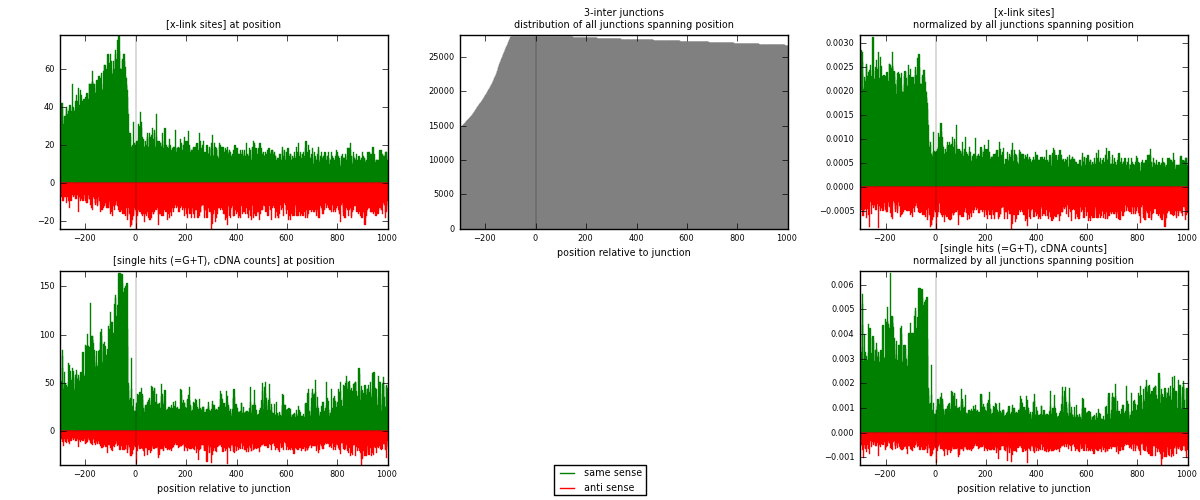 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 26259 | 1.06% | 8.16% | 65.09% | 6699 | 2.66076 |
| anti | 14083 | 0.57% | 4.38% | 34.91% | 3935 | 1.42699 |
| both | 40342 | 1.63% | 12.54% | 100.00% | 9869 | 4.08775 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 43620 | 1.54% | 11.04% | 73.08% | 6699 | 4.4199 |
| anti | 16072 | 0.57% | 4.07% | 26.92% | 3935 | 1.62853 |
| both | 59692 | 2.10% | 15.11% | 100.00% | 9869 | 6.04843 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
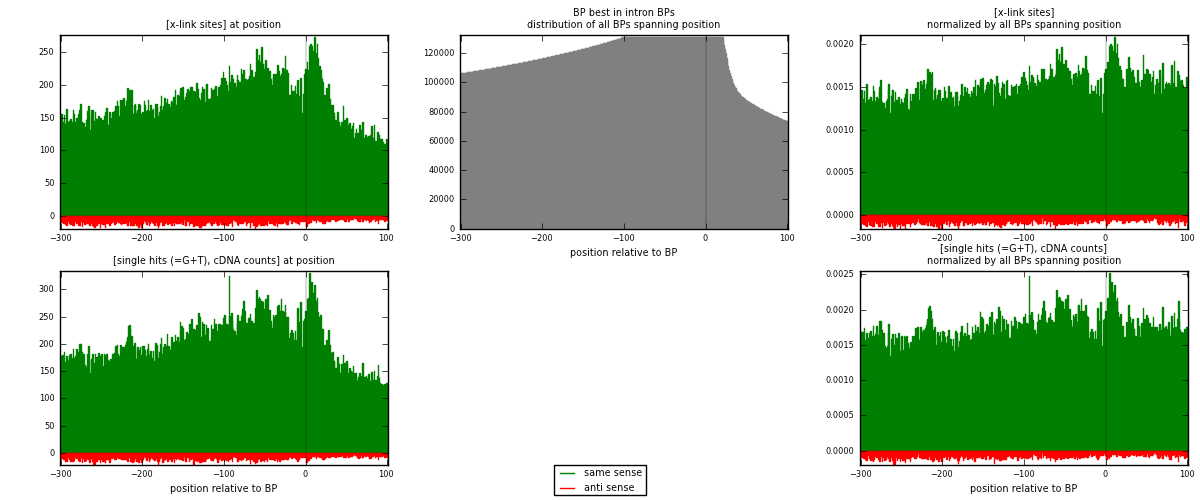 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 69360 | 2.80% | 21.57% | 95.26% | 29081 | 2.23728 |
| anti | 3452 | 0.14% | 1.07% | 4.74% | 2284 | 0.111348 |
| both | 72812 | 2.94% | 22.64% | 100.00% | 31002 | 2.34862 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 80503 | 2.84% | 20.38% | 95.63% | 29081 | 2.5967 |
| anti | 3680 | 0.13% | 0.93% | 4.37% | 2284 | 0.118702 |
| both | 84183 | 2.97% | 21.31% | 100.00% | 31002 | 2.71541 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.