RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
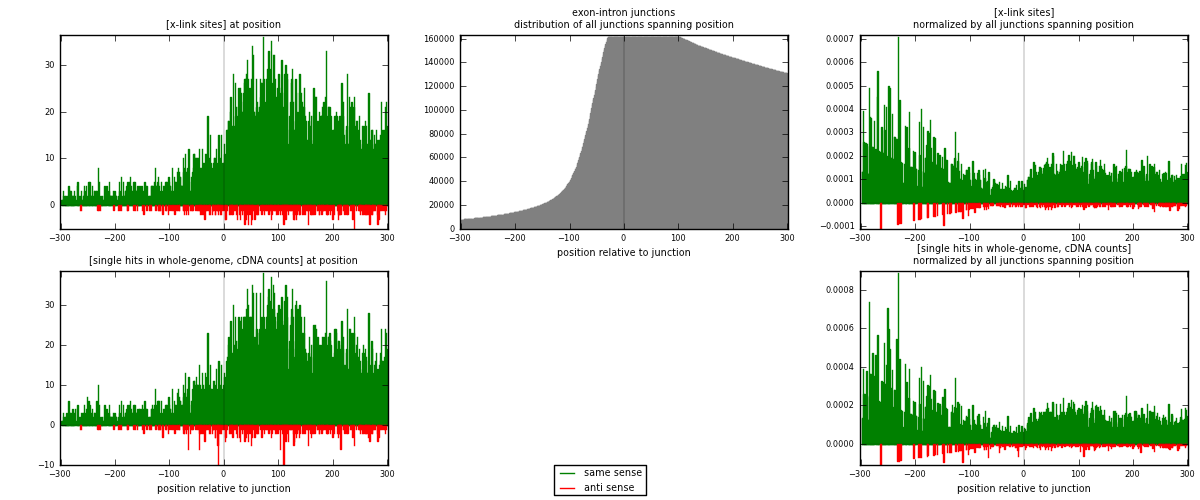 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6834 | 4.19% | 27.99% | 94.26% | 5027 | 1.27357 |
| anti | 416 | 0.26% | 1.70% | 5.74% | 352 | 0.0775252 |
| both | 7250 | 4.44% | 29.70% | 100.00% | 5366 | 1.3511 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 7364 | 4.05% | 24.37% | 93.88% | 5027 | 1.37234 |
| anti | 480 | 0.26% | 1.59% | 6.12% | 352 | 0.0894521 |
| both | 7844 | 4.31% | 25.96% | 100.00% | 5366 | 1.4618 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
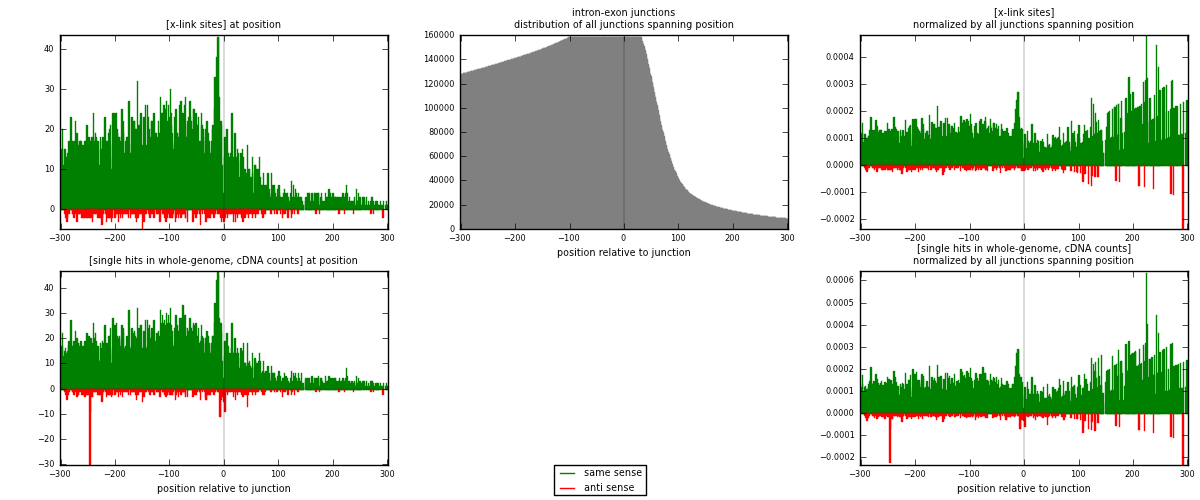 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6520 | 4.00% | 26.71% | 94.60% | 5008 | 1.22556 |
| anti | 372 | 0.23% | 1.52% | 5.40% | 323 | 0.0699248 |
| both | 6892 | 4.22% | 28.23% | 100.00% | 5320 | 1.29549 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 7027 | 3.87% | 23.26% | 93.68% | 5008 | 1.32086 |
| anti | 474 | 0.26% | 1.57% | 6.32% | 323 | 0.0890977 |
| both | 7501 | 4.13% | 24.83% | 100.00% | 5320 | 1.40996 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
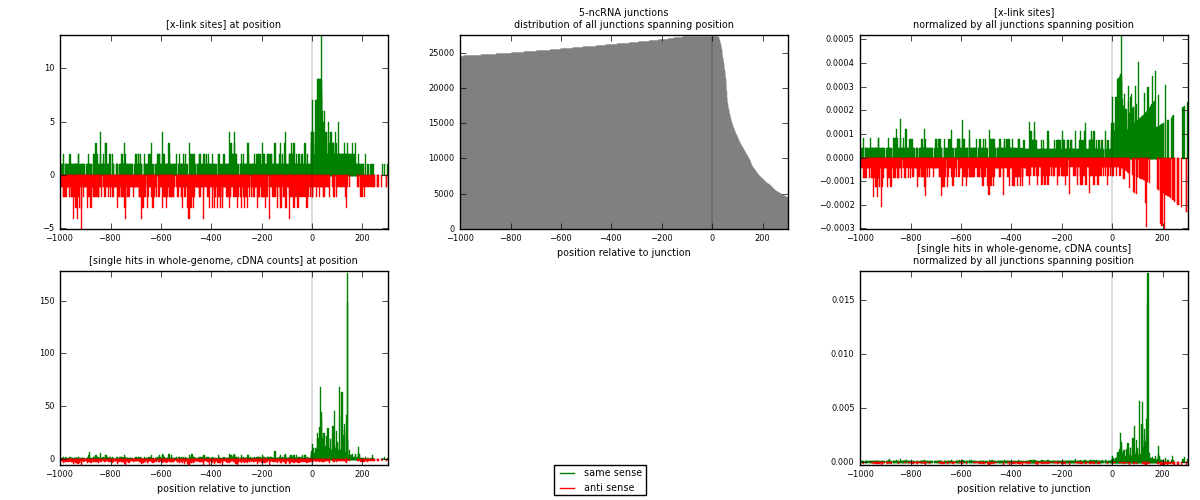 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1122 | 0.69% | 4.60% | 54.55% | 526 | 0.981627 |
| anti | 935 | 0.57% | 3.83% | 45.45% | 625 | 0.818023 |
| both | 2057 | 1.26% | 8.43% | 100.00% | 1143 | 1.79965 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2657 | 1.46% | 8.79% | 72.58% | 526 | 2.32458 |
| anti | 1004 | 0.55% | 3.32% | 27.42% | 625 | 0.87839 |
| both | 3661 | 2.01% | 12.12% | 100.00% | 1143 | 3.20297 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
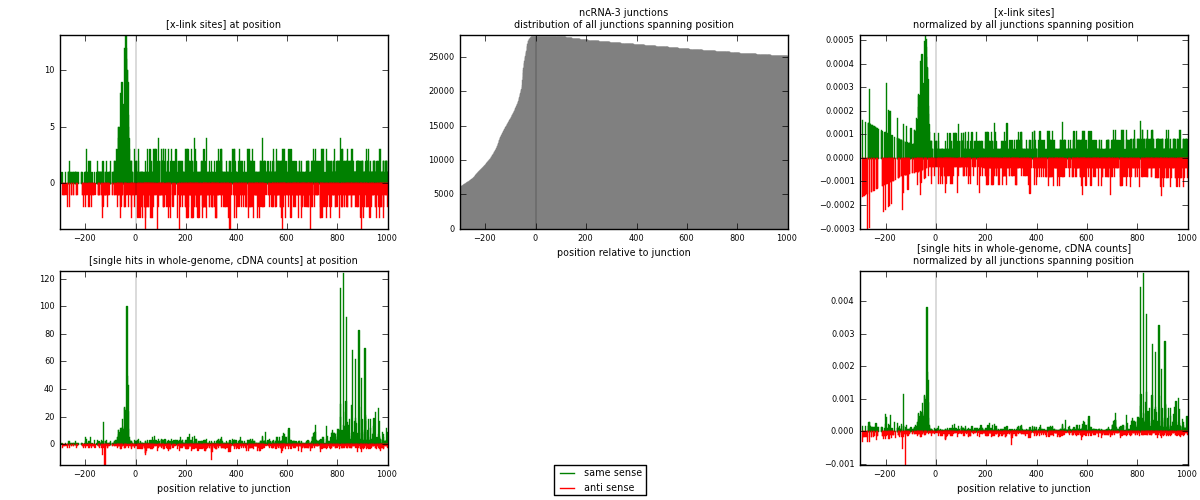 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1405 | 0.86% | 5.75% | 59.28% | 584 | 1.18967 |
| anti | 965 | 0.59% | 3.95% | 40.72% | 608 | 0.817104 |
| both | 2370 | 1.45% | 9.71% | 100.00% | 1181 | 2.00677 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3680 | 2.02% | 12.18% | 76.91% | 584 | 3.116 |
| anti | 1105 | 0.61% | 3.66% | 23.09% | 608 | 0.935648 |
| both | 4785 | 2.63% | 15.84% | 100.00% | 1181 | 4.05165 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
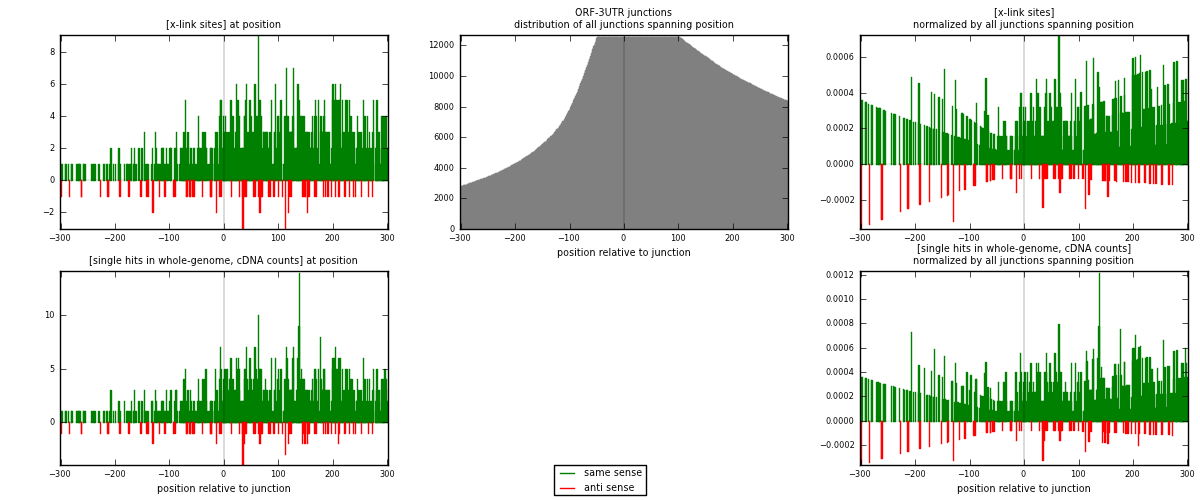 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 955 | 0.59% | 3.91% | 91.56% | 590 | 1.43825 |
| anti | 88 | 0.05% | 0.36% | 8.44% | 75 | 0.13253 |
| both | 1043 | 0.64% | 4.27% | 100.00% | 664 | 1.57078 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1047 | 0.58% | 3.47% | 91.84% | 590 | 1.57681 |
| anti | 93 | 0.05% | 0.31% | 8.16% | 75 | 0.14006 |
| both | 1140 | 0.63% | 3.77% | 100.00% | 664 | 1.71687 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
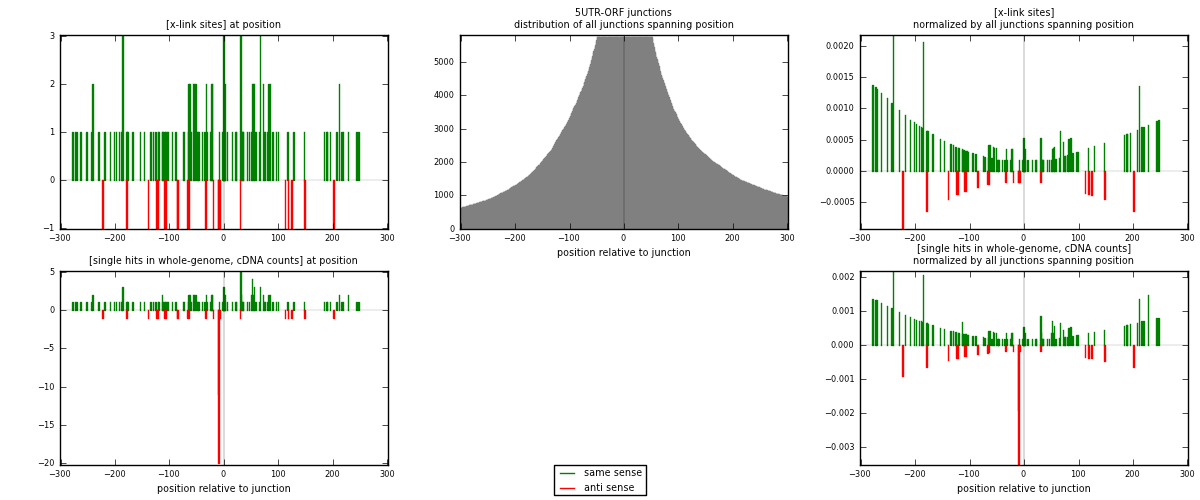 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 130 | 0.08% | 0.53% | 84.97% | 84 | 1.28713 |
| anti | 23 | 0.01% | 0.09% | 15.03% | 17 | 0.227723 |
| both | 153 | 0.09% | 0.63% | 100.00% | 101 | 1.51485 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 139 | 0.08% | 0.46% | 72.77% | 84 | 1.37624 |
| anti | 52 | 0.03% | 0.17% | 27.23% | 17 | 0.514851 |
| both | 191 | 0.11% | 0.63% | 100.00% | 101 | 1.89109 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
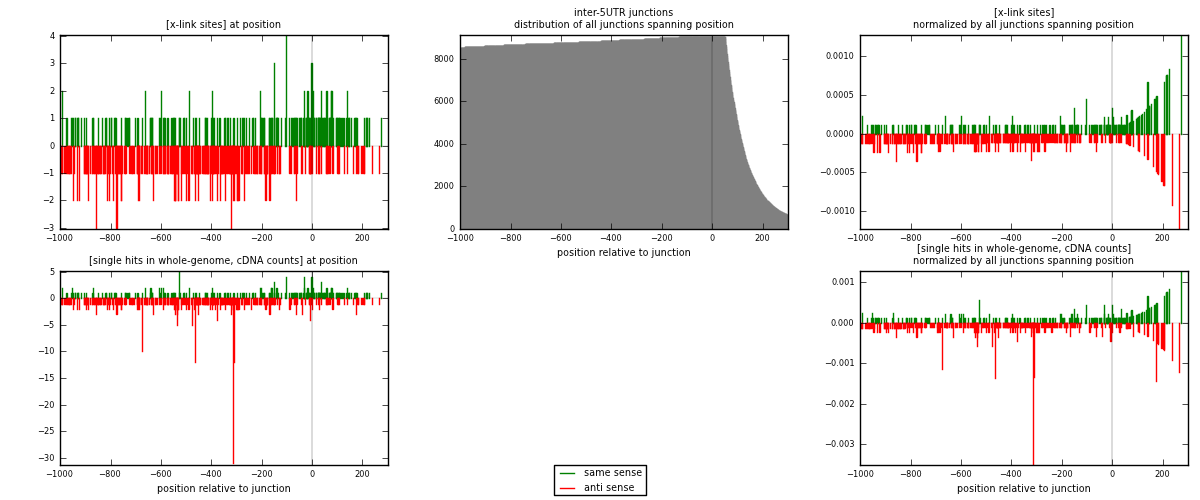 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 216 | 0.13% | 0.88% | 38.23% | 141 | 0.629738 |
| anti | 349 | 0.21% | 1.43% | 61.77% | 206 | 1.01749 |
| both | 565 | 0.35% | 2.31% | 100.00% | 343 | 1.64723 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 234 | 0.13% | 0.77% | 34.36% | 141 | 0.682216 |
| anti | 447 | 0.25% | 1.48% | 65.64% | 206 | 1.30321 |
| both | 681 | 0.37% | 2.25% | 100.00% | 343 | 1.98542 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
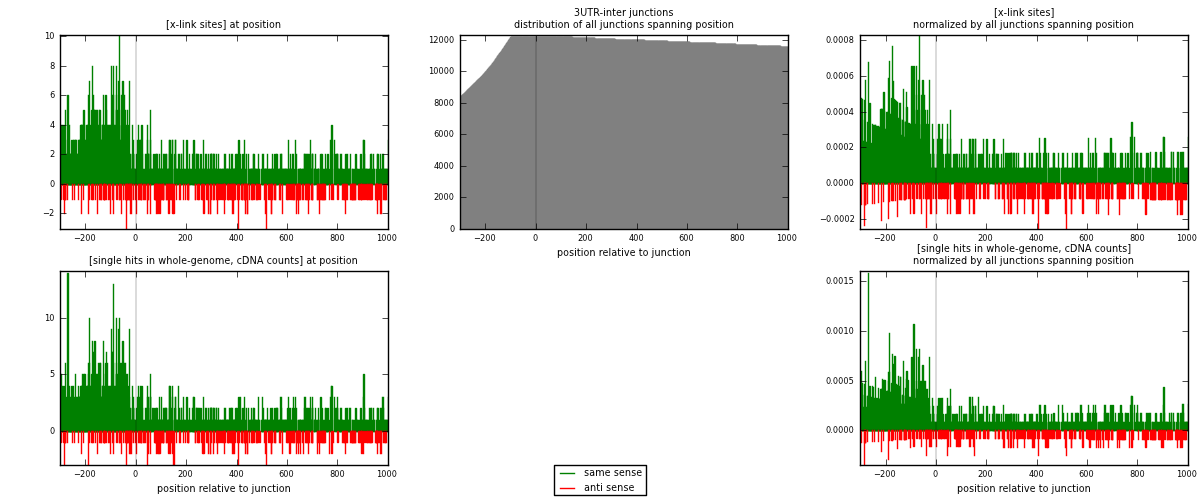 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1461 | 0.90% | 5.98% | 80.85% | 900 | 1.29292 |
| anti | 346 | 0.21% | 1.42% | 19.15% | 239 | 0.306195 |
| both | 1807 | 1.11% | 7.40% | 100.00% | 1130 | 1.59912 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1592 | 0.88% | 5.27% | 81.35% | 900 | 1.40885 |
| anti | 365 | 0.20% | 1.21% | 18.65% | 239 | 0.323009 |
| both | 1957 | 1.08% | 6.48% | 100.00% | 1130 | 1.73186 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
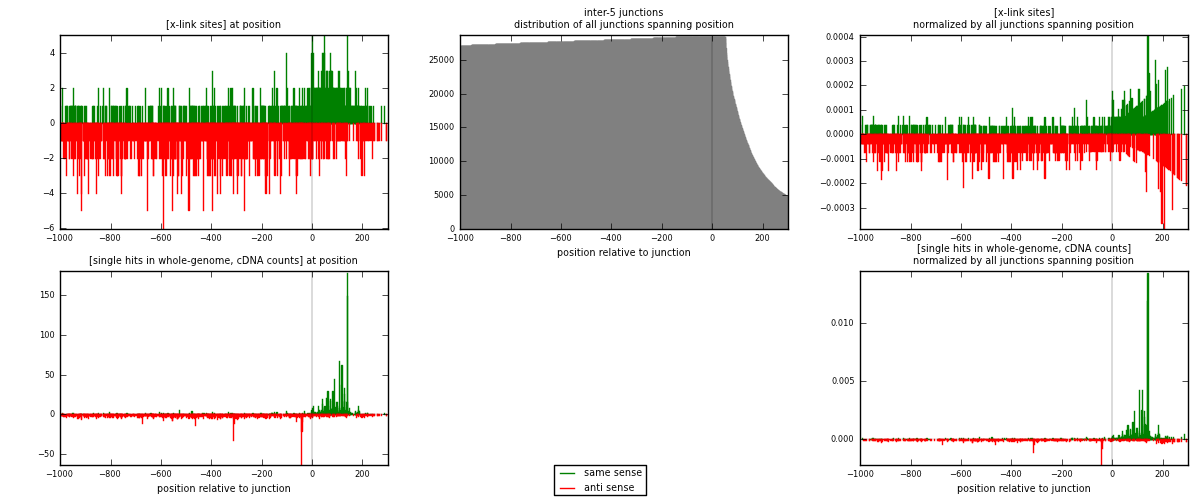
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 567 | 0.35% | 2.32% | 33.29% | 323 | 0.553171 |
| anti | 1136 | 0.70% | 4.65% | 66.71% | 711 | 1.10829 |
| both | 1703 | 1.04% | 6.98% | 100.00% | 1025 | 1.66146 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1658 | 0.91% | 5.49% | 54.27% | 323 | 1.61756 |
| anti | 1397 | 0.77% | 4.62% | 45.73% | 711 | 1.36293 |
| both | 3055 | 1.68% | 10.11% | 100.00% | 1025 | 2.98049 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
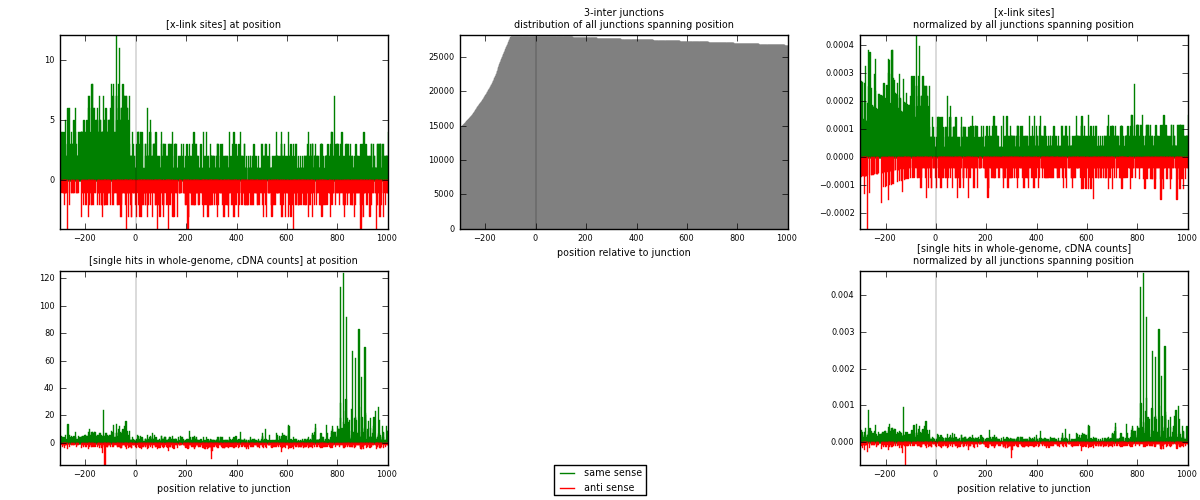
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2228 | 1.37% | 9.13% | 69.39% | 1120 | 1.28637 |
| anti | 983 | 0.60% | 4.03% | 30.61% | 625 | 0.567552 |
| both | 3211 | 1.97% | 13.15% | 100.00% | 1732 | 1.85393 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4260 | 2.34% | 14.10% | 79.55% | 1120 | 2.45958 |
| anti | 1095 | 0.60% | 3.62% | 20.45% | 625 | 0.632217 |
| both | 5355 | 2.95% | 17.72% | 100.00% | 1732 | 3.0918 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
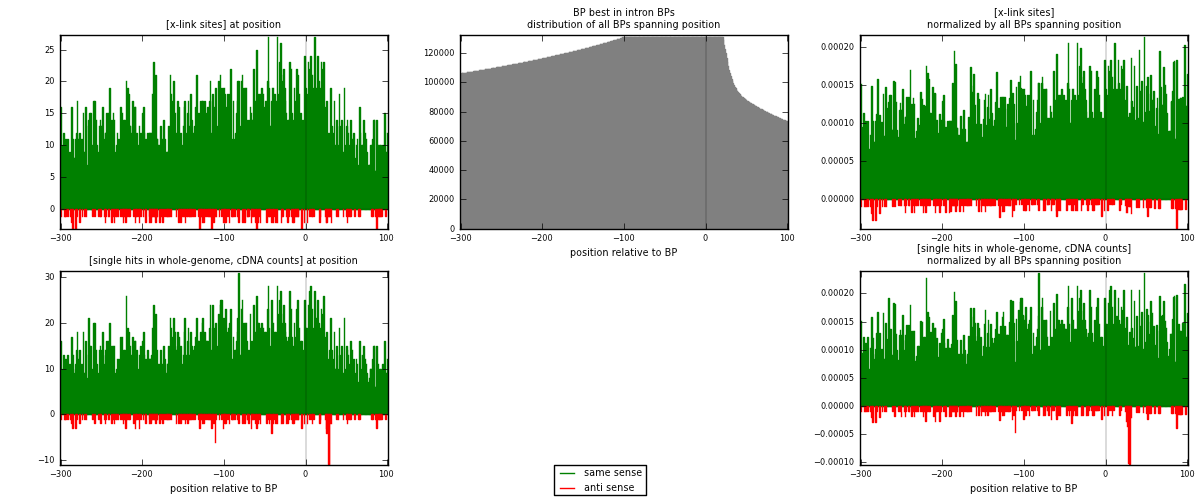
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5372 | 3.29% | 22.00% | 95.47% | 3925 | 1.29477 |
| anti | 255 | 0.16% | 1.04% | 4.53% | 232 | 0.0614606 |
| both | 5627 | 3.45% | 23.05% | 100.00% | 4149 | 1.35623 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 5838 | 3.21% | 19.32% | 95.10% | 3925 | 1.40709 |
| anti | 301 | 0.17% | 1.00% | 4.90% | 232 | 0.0725476 |
| both | 6139 | 3.38% | 20.32% | 100.00% | 4149 | 1.47963 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.