RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
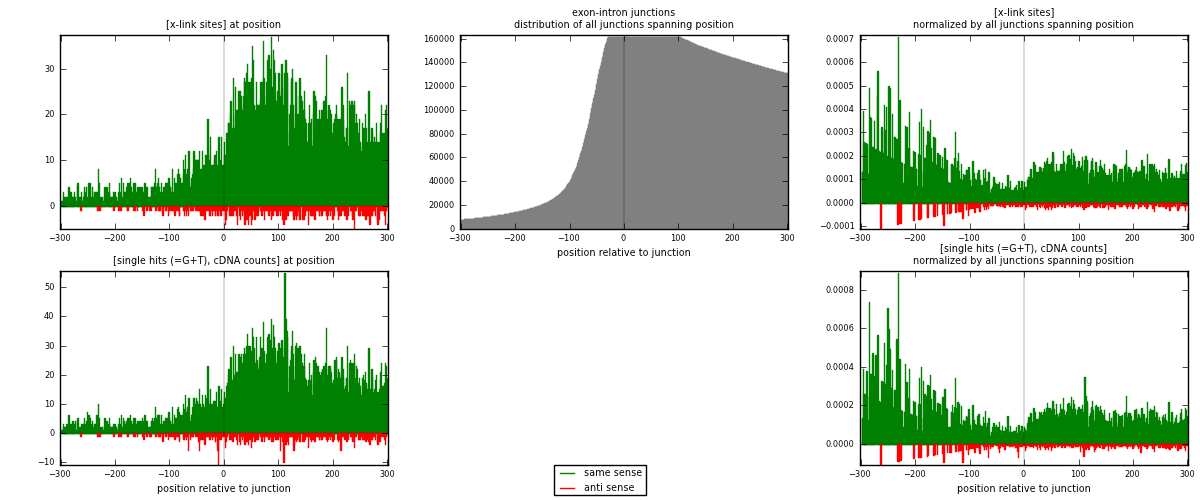 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6987 | 4.10% | 27.68% | 93.76% | 5120 | 1.2699 |
| anti | 465 | 0.27% | 1.84% | 6.24% | 396 | 0.0845147 |
| both | 7452 | 4.37% | 29.52% | 100.00% | 5502 | 1.35442 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 7579 | 3.97% | 24.15% | 93.38% | 5120 | 1.3775 |
| anti | 537 | 0.28% | 1.71% | 6.62% | 396 | 0.0976009 |
| both | 8116 | 4.26% | 25.86% | 100.00% | 5502 | 1.4751 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
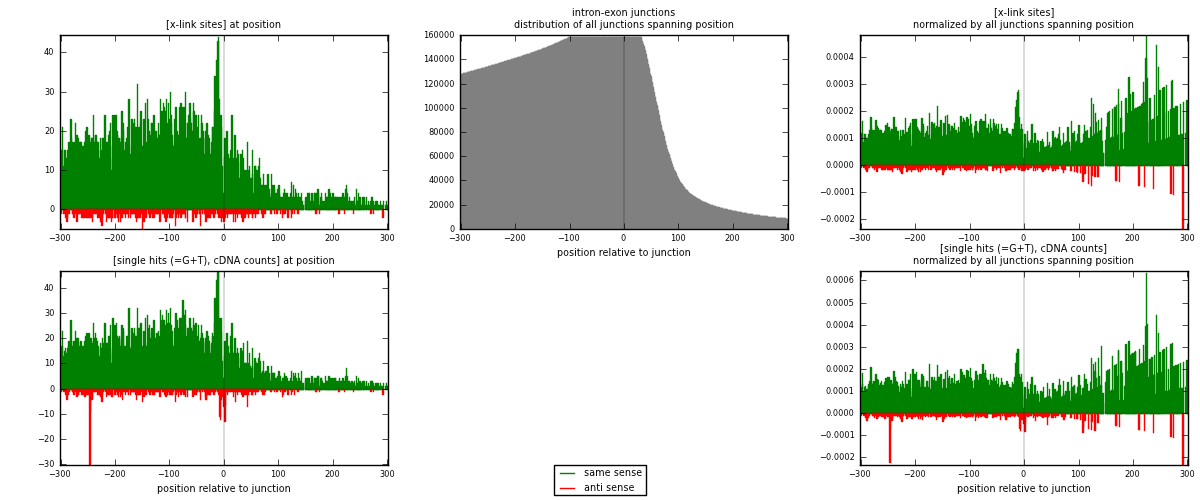 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6653 | 3.90% | 26.35% | 94.29% | 5098 | 1.2241 |
| anti | 403 | 0.24% | 1.60% | 5.71% | 351 | 0.074149 |
| both | 7056 | 4.14% | 27.95% | 100.00% | 5435 | 1.29825 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 7185 | 3.77% | 22.89% | 93.20% | 5098 | 1.32199 |
| anti | 524 | 0.27% | 1.67% | 6.80% | 351 | 0.0964121 |
| both | 7709 | 4.04% | 24.56% | 100.00% | 5435 | 1.4184 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
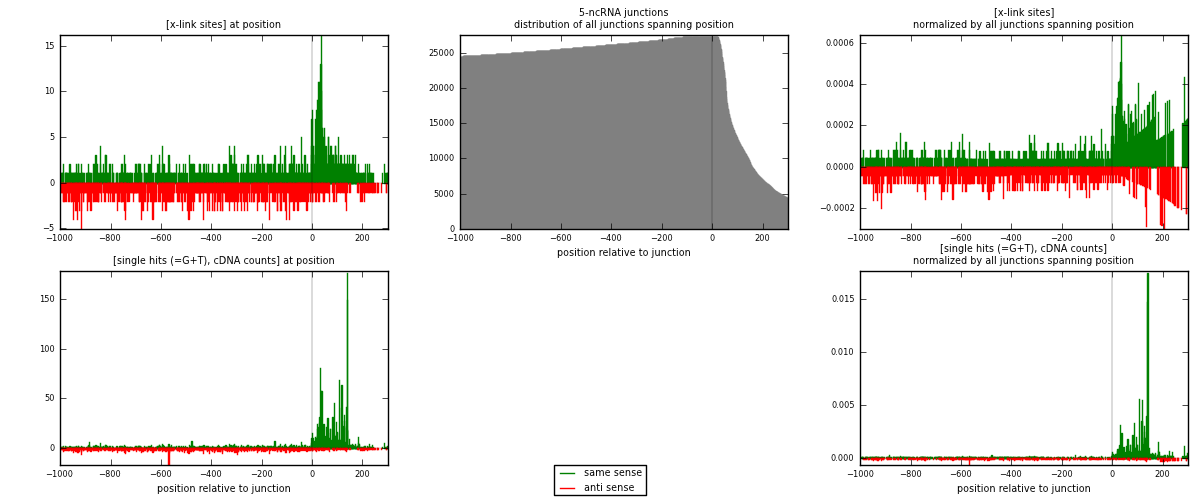 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1233 | 0.72% | 4.88% | 55.57% | 572 | 1.01148 |
| anti | 986 | 0.58% | 3.91% | 44.43% | 659 | 0.80886 |
| both | 2219 | 1.30% | 8.79% | 100.00% | 1219 | 1.82034 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2886 | 1.51% | 9.19% | 72.81% | 572 | 2.36751 |
| anti | 1078 | 0.57% | 3.43% | 27.19% | 659 | 0.884331 |
| both | 3964 | 2.08% | 12.63% | 100.00% | 1219 | 3.25185 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
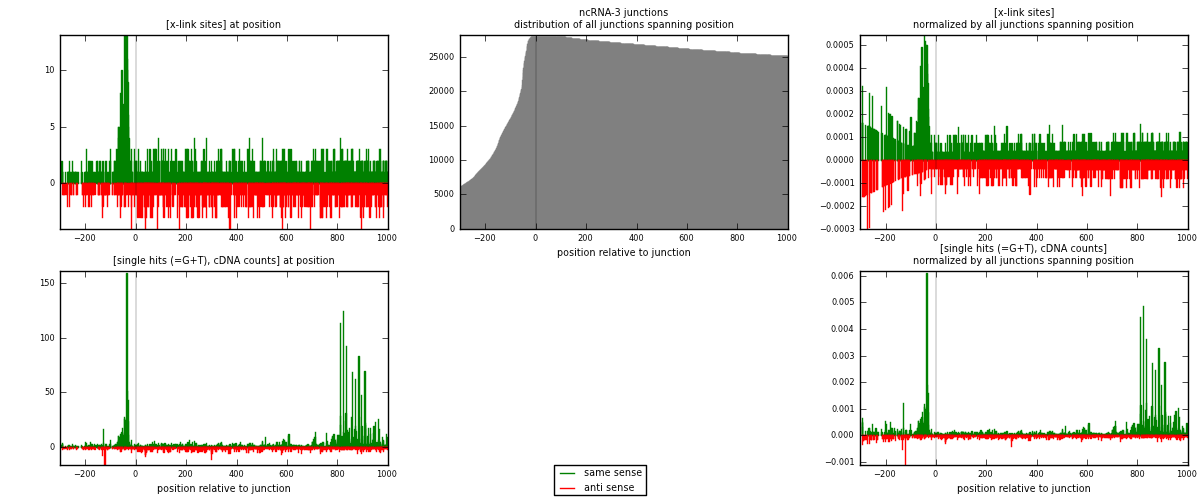 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1472 | 0.86% | 5.83% | 59.07% | 615 | 1.18519 |
| anti | 1020 | 0.60% | 4.04% | 40.93% | 639 | 0.821256 |
| both | 2492 | 1.46% | 9.87% | 100.00% | 1242 | 2.00644 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3836 | 2.01% | 12.22% | 76.69% | 615 | 3.08857 |
| anti | 1166 | 0.61% | 3.71% | 23.31% | 639 | 0.938808 |
| both | 5002 | 2.62% | 15.94% | 100.00% | 1242 | 4.02738 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
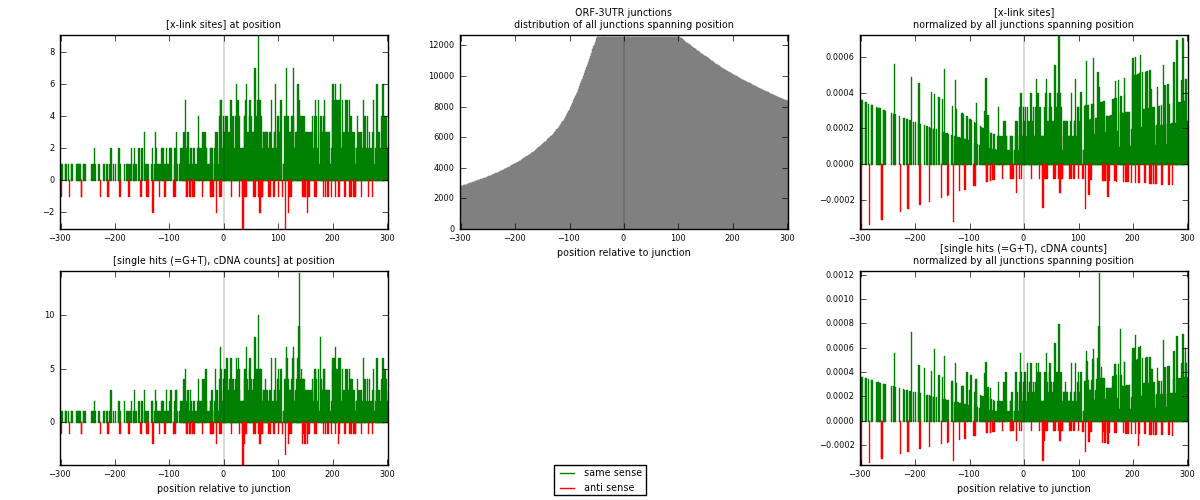
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 982 | 0.58% | 3.89% | 91.43% | 606 | 1.43988 |
| anti | 92 | 0.05% | 0.36% | 8.57% | 77 | 0.134897 |
| both | 1074 | 0.63% | 4.25% | 100.00% | 682 | 1.57478 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1076 | 0.56% | 3.43% | 91.73% | 606 | 1.57771 |
| anti | 97 | 0.05% | 0.31% | 8.27% | 77 | 0.142229 |
| both | 1173 | 0.62% | 3.74% | 100.00% | 682 | 1.71994 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
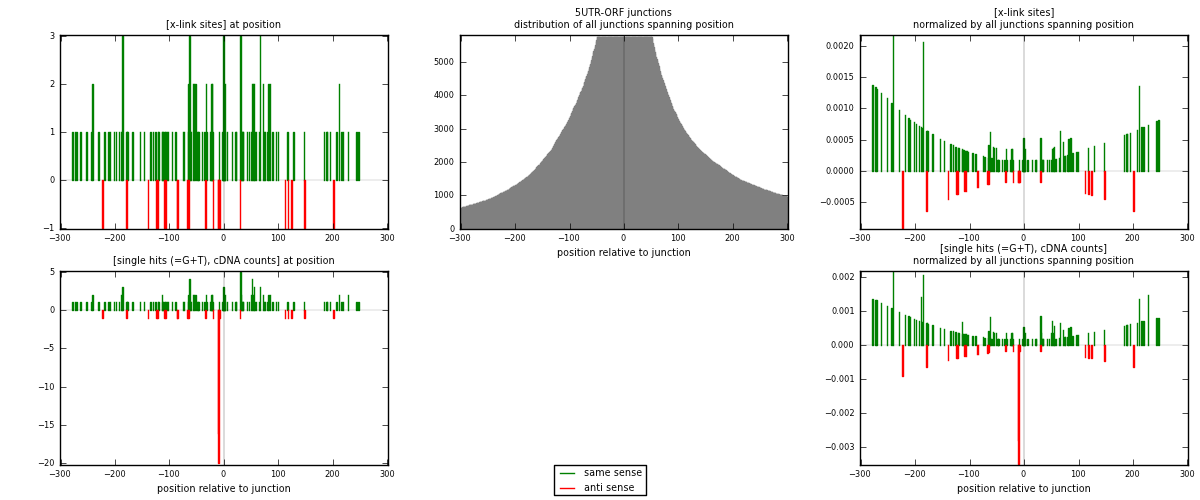
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 134 | 0.08% | 0.53% | 85.35% | 85 | 1.31373 |
| anti | 23 | 0.01% | 0.09% | 14.65% | 17 | 0.22549 |
| both | 157 | 0.09% | 0.62% | 100.00% | 102 | 1.53922 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 145 | 0.08% | 0.46% | 71.78% | 85 | 1.42157 |
| anti | 57 | 0.03% | 0.18% | 28.22% | 17 | 0.558824 |
| both | 202 | 0.11% | 0.64% | 100.00% | 102 | 1.98039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
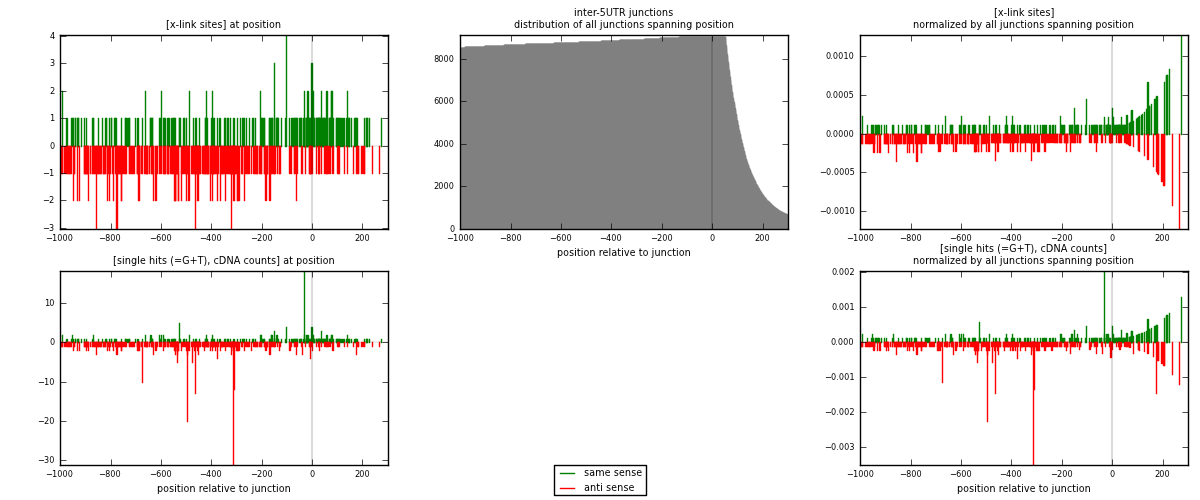
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 221 | 0.13% | 0.88% | 38.04% | 144 | 0.626062 |
| anti | 360 | 0.21% | 1.43% | 61.96% | 213 | 1.01983 |
| both | 581 | 0.34% | 2.30% | 100.00% | 353 | 1.64589 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 254 | 0.13% | 0.81% | 34.60% | 144 | 0.719547 |
| anti | 480 | 0.25% | 1.53% | 65.40% | 213 | 1.35977 |
| both | 734 | 0.38% | 2.34% | 100.00% | 353 | 2.07932 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
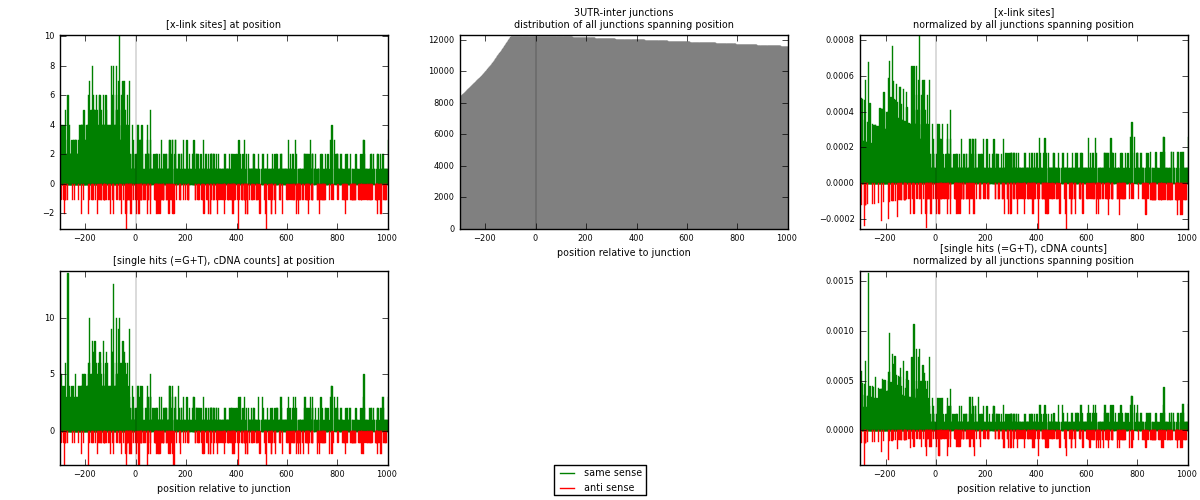
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1500 | 0.88% | 5.94% | 80.73% | 924 | 1.29199 |
| anti | 358 | 0.21% | 1.42% | 19.27% | 247 | 0.308355 |
| both | 1858 | 1.09% | 7.36% | 100.00% | 1161 | 1.60034 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1631 | 0.86% | 5.20% | 81.10% | 924 | 1.40482 |
| anti | 380 | 0.20% | 1.21% | 18.90% | 247 | 0.327304 |
| both | 2011 | 1.05% | 6.41% | 100.00% | 1161 | 1.73213 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
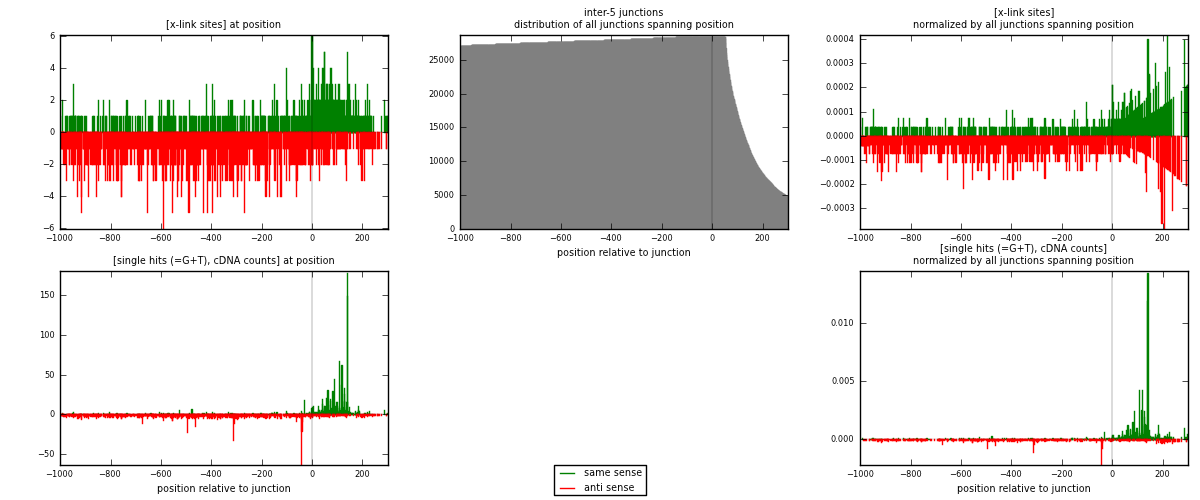
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 627 | 0.37% | 2.48% | 34.74% | 341 | 0.586529 |
| anti | 1178 | 0.69% | 4.67% | 65.26% | 738 | 1.10196 |
| both | 1805 | 1.06% | 7.15% | 100.00% | 1069 | 1.68849 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1749 | 0.92% | 5.57% | 54.45% | 341 | 1.63611 |
| anti | 1463 | 0.77% | 4.66% | 45.55% | 738 | 1.36857 |
| both | 3212 | 1.68% | 10.23% | 100.00% | 1069 | 3.00468 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
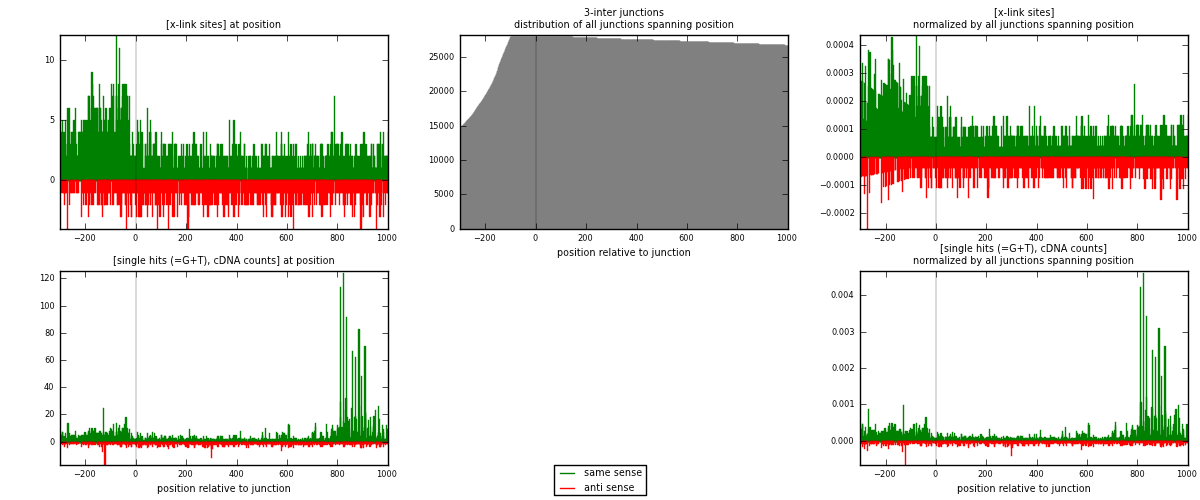
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2309 | 1.35% | 9.15% | 69.07% | 1160 | 1.28635 |
| anti | 1034 | 0.61% | 4.10% | 30.93% | 650 | 0.576045 |
| both | 3343 | 1.96% | 13.24% | 100.00% | 1795 | 1.8624 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4370 | 2.29% | 13.92% | 79.14% | 1160 | 2.43454 |
| anti | 1152 | 0.60% | 3.67% | 20.86% | 650 | 0.641783 |
| both | 5522 | 2.90% | 17.59% | 100.00% | 1795 | 3.07632 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
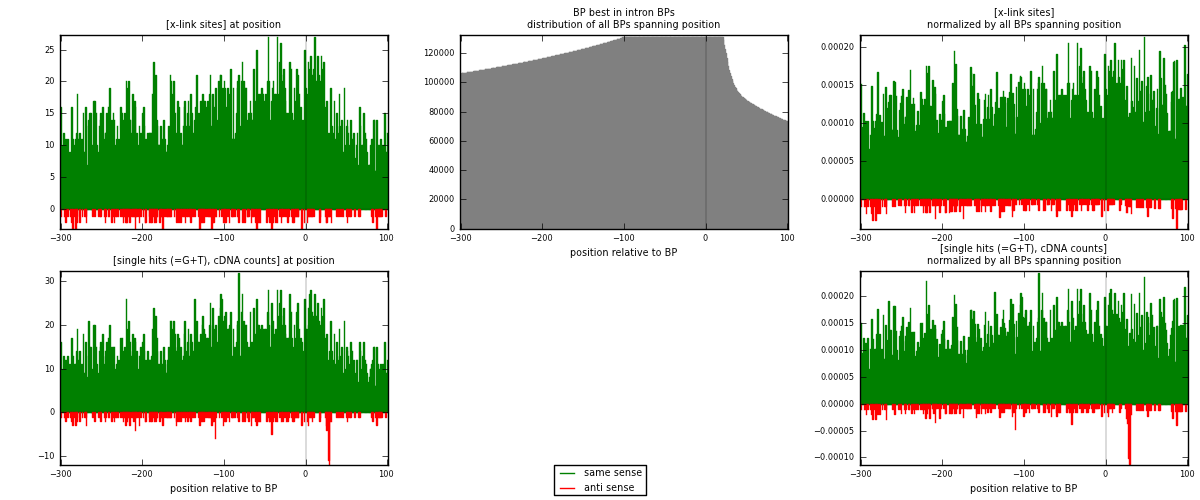
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5481 | 3.21% | 21.71% | 94.91% | 3995 | 1.28904 |
| anti | 294 | 0.17% | 1.16% | 5.09% | 267 | 0.0691439 |
| both | 5775 | 3.38% | 22.88% | 100.00% | 4252 | 1.35818 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 5963 | 3.13% | 19.00% | 94.52% | 3995 | 1.4024 |
| anti | 346 | 0.18% | 1.10% | 5.48% | 267 | 0.0813735 |
| both | 6309 | 3.31% | 20.10% | 100.00% | 4252 | 1.48377 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.