RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
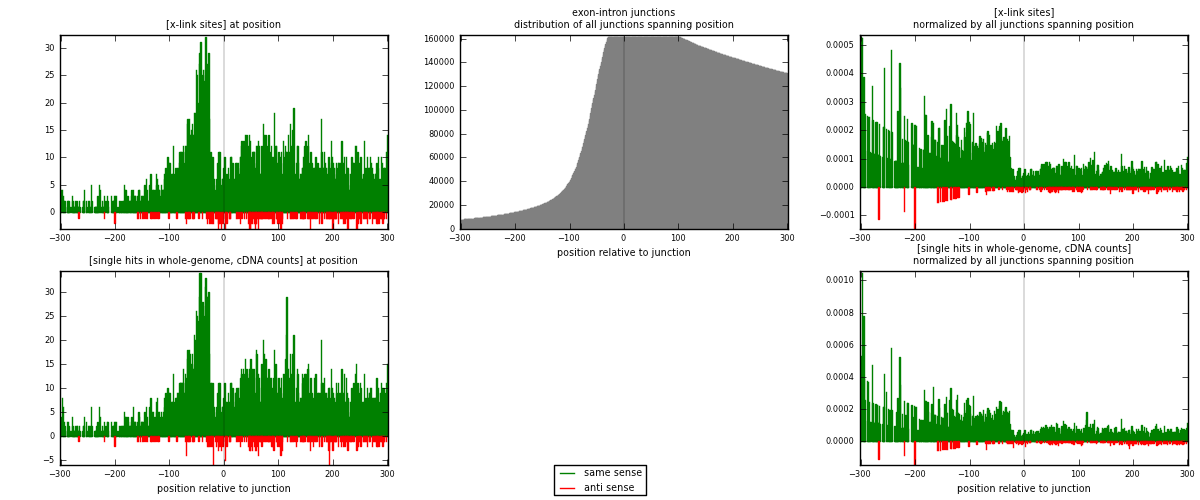 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4184 | 4.76% | 23.99% | 94.13% | 3273 | 1.20611 |
| anti | 261 | 0.30% | 1.50% | 5.87% | 198 | 0.0752378 |
| both | 4445 | 5.05% | 25.48% | 100.00% | 3469 | 1.28135 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4577 | 4.62% | 21.51% | 94.02% | 3273 | 1.3194 |
| anti | 291 | 0.29% | 1.37% | 5.98% | 198 | 0.0838858 |
| both | 4868 | 4.92% | 22.88% | 100.00% | 3469 | 1.40329 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
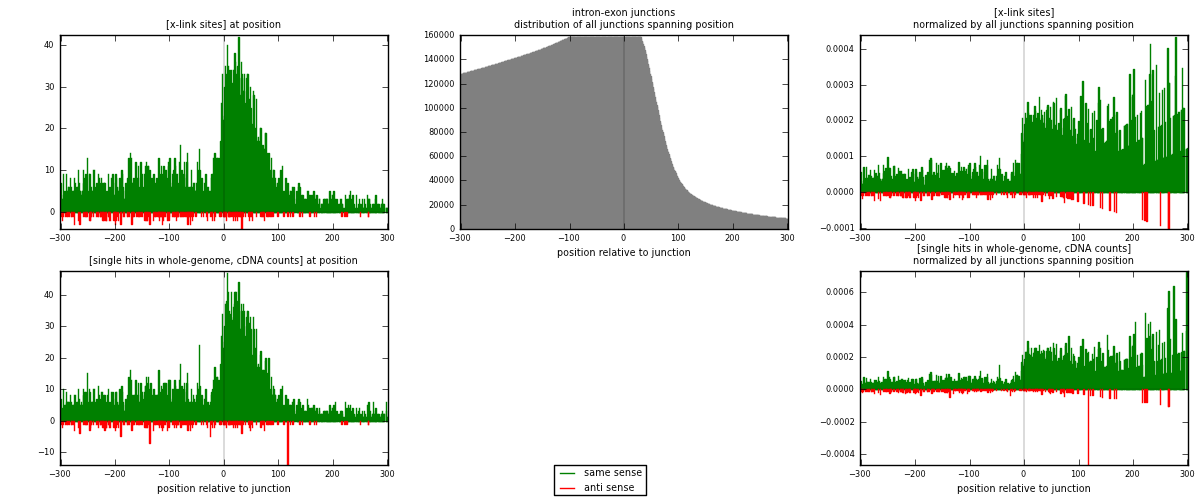 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4766 | 5.42% | 27.32% | 95.19% | 3851 | 1.18087 |
| anti | 241 | 0.27% | 1.38% | 4.81% | 192 | 0.0597126 |
| both | 5007 | 5.69% | 28.71% | 100.00% | 4036 | 1.24058 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 5144 | 5.20% | 24.18% | 94.89% | 3851 | 1.27453 |
| anti | 277 | 0.28% | 1.30% | 5.11% | 192 | 0.0686323 |
| both | 5421 | 5.48% | 25.48% | 100.00% | 4036 | 1.34316 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
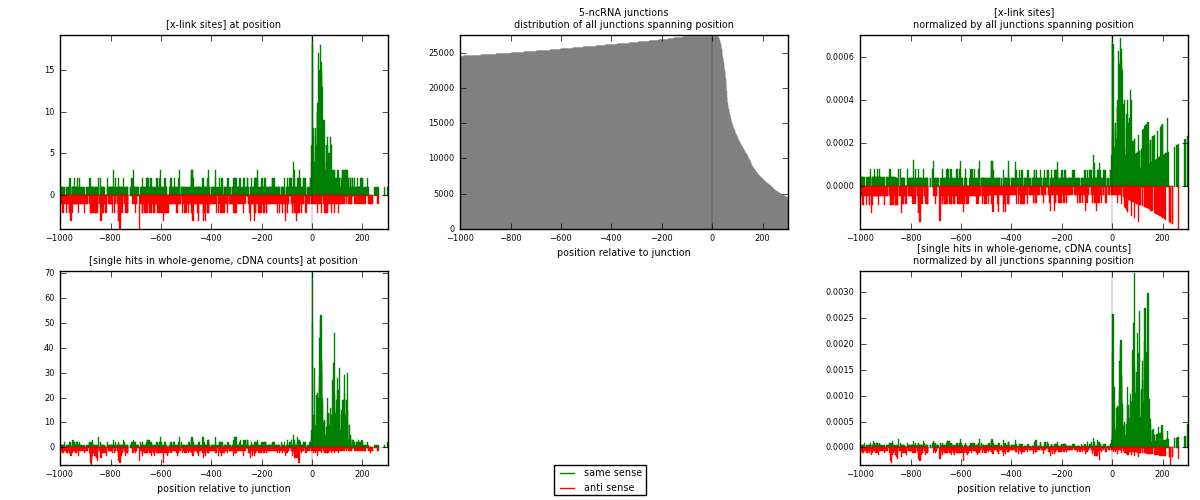 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1192 | 1.36% | 6.83% | 64.29% | 541 | 1.24296 |
| anti | 662 | 0.75% | 3.80% | 35.71% | 426 | 0.690302 |
| both | 1854 | 2.11% | 10.63% | 100.00% | 959 | 1.93326 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2575 | 2.60% | 12.10% | 77.37% | 541 | 2.68509 |
| anti | 753 | 0.76% | 3.54% | 22.63% | 426 | 0.785193 |
| both | 3328 | 3.36% | 15.64% | 100.00% | 959 | 3.47028 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
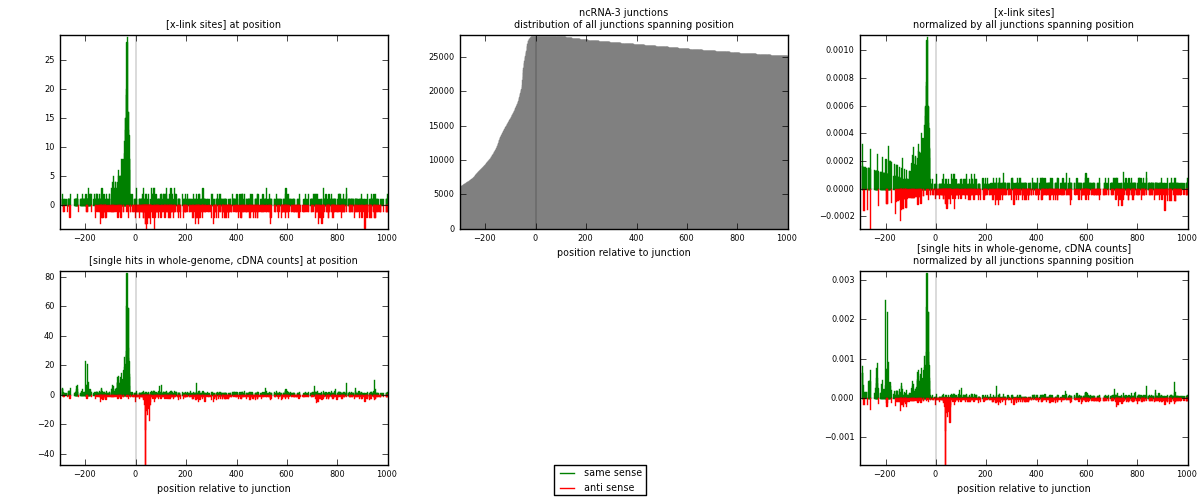 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1108 | 1.26% | 6.35% | 65.91% | 483 | 1.39021 |
| anti | 573 | 0.65% | 3.29% | 34.09% | 319 | 0.718946 |
| both | 1681 | 1.91% | 9.64% | 100.00% | 797 | 2.10916 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1825 | 1.84% | 8.58% | 69.15% | 483 | 2.28984 |
| anti | 814 | 0.82% | 3.83% | 30.85% | 319 | 1.02133 |
| both | 2639 | 2.67% | 12.40% | 100.00% | 797 | 3.31117 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
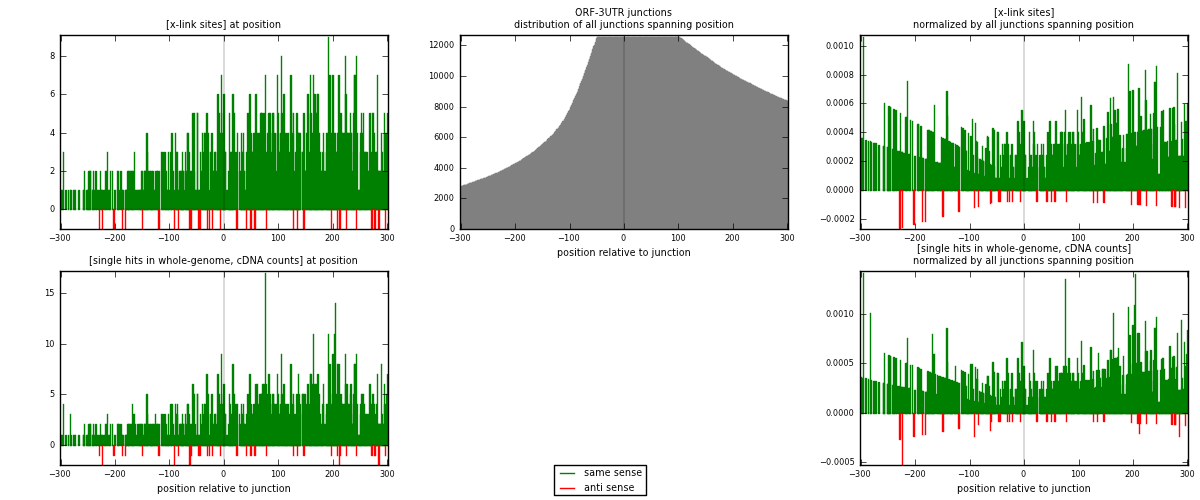 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1300 | 1.48% | 7.45% | 97.31% | 839 | 1.4977 |
| anti | 36 | 0.04% | 0.21% | 2.69% | 30 | 0.0414747 |
| both | 1336 | 1.52% | 7.66% | 100.00% | 868 | 1.53917 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1466 | 1.48% | 6.89% | 97.28% | 839 | 1.68894 |
| anti | 41 | 0.04% | 0.19% | 2.72% | 30 | 0.047235 |
| both | 1507 | 1.52% | 7.08% | 100.00% | 868 | 1.73618 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
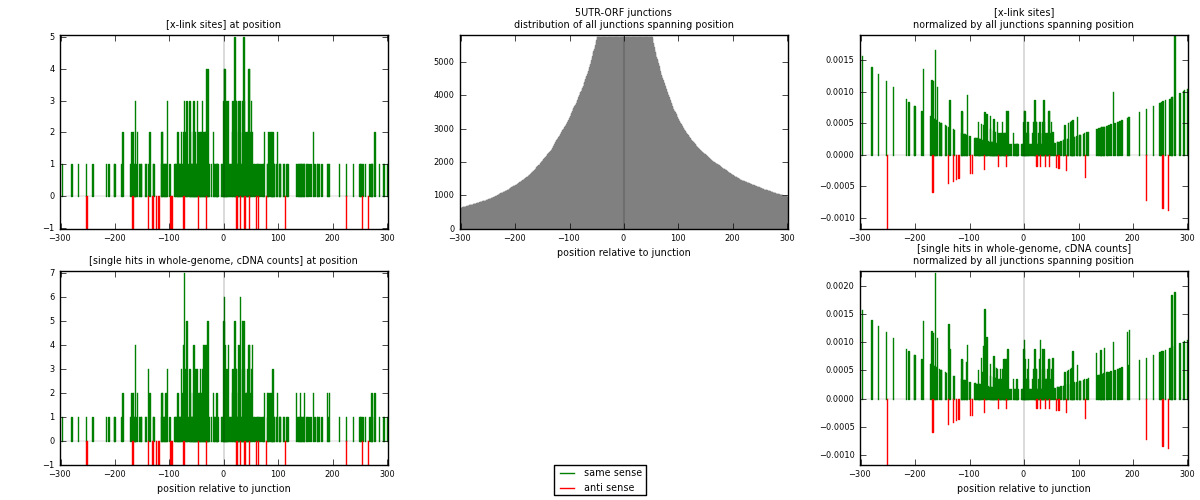 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 324 | 0.37% | 1.86% | 93.64% | 216 | 1.38462 |
| anti | 22 | 0.03% | 0.13% | 6.36% | 20 | 0.0940171 |
| both | 346 | 0.39% | 1.98% | 100.00% | 234 | 1.47863 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 380 | 0.38% | 1.79% | 94.53% | 216 | 1.62393 |
| anti | 22 | 0.02% | 0.10% | 5.47% | 20 | 0.0940171 |
| both | 402 | 0.41% | 1.89% | 100.00% | 234 | 1.71795 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
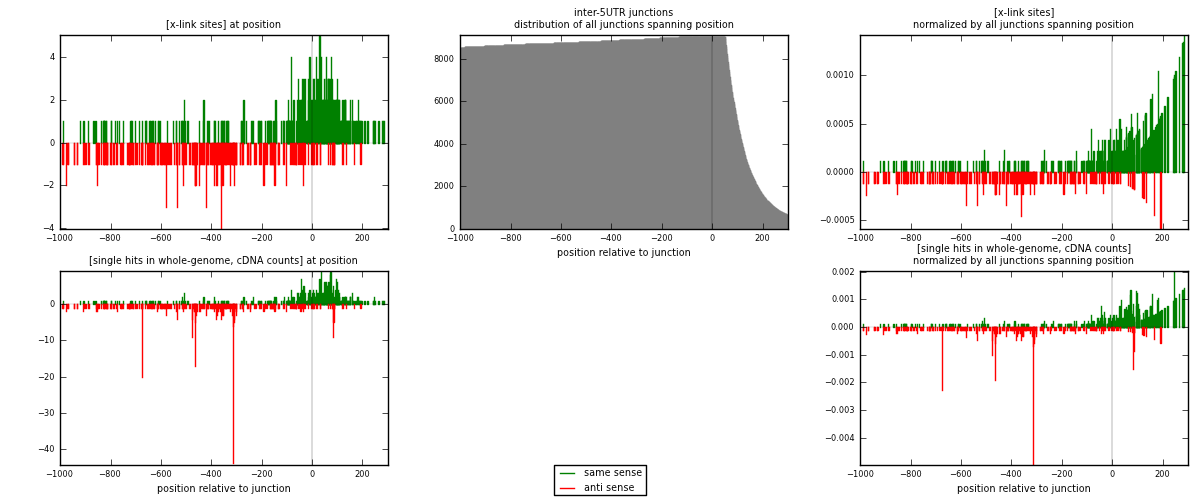 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 412 | 0.47% | 2.36% | 60.23% | 213 | 1.09867 |
| anti | 272 | 0.31% | 1.56% | 39.77% | 162 | 0.725333 |
| both | 684 | 0.78% | 3.92% | 100.00% | 375 | 1.824 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 519 | 0.52% | 2.44% | 55.21% | 213 | 1.384 |
| anti | 421 | 0.43% | 1.98% | 44.79% | 162 | 1.12267 |
| both | 940 | 0.95% | 4.42% | 100.00% | 375 | 2.50667 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
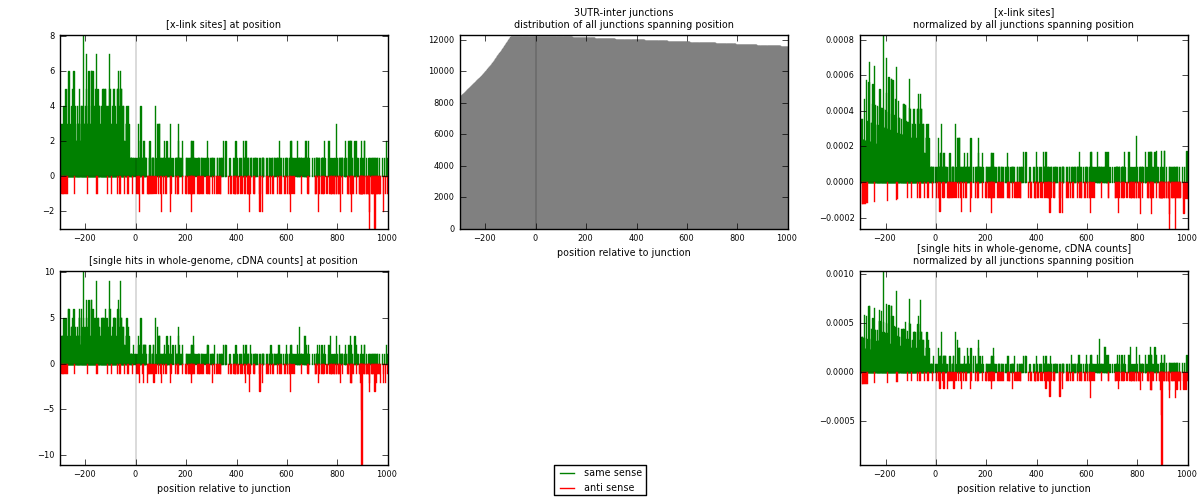 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1076 | 1.22% | 6.17% | 83.15% | 749 | 1.19158 |
| anti | 218 | 0.25% | 1.25% | 16.85% | 163 | 0.241417 |
| both | 1294 | 1.47% | 7.42% | 100.00% | 903 | 1.433 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1183 | 1.19% | 5.56% | 82.73% | 749 | 1.31008 |
| anti | 247 | 0.25% | 1.16% | 17.27% | 163 | 0.273533 |
| both | 1430 | 1.44% | 6.72% | 100.00% | 903 | 1.58361 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
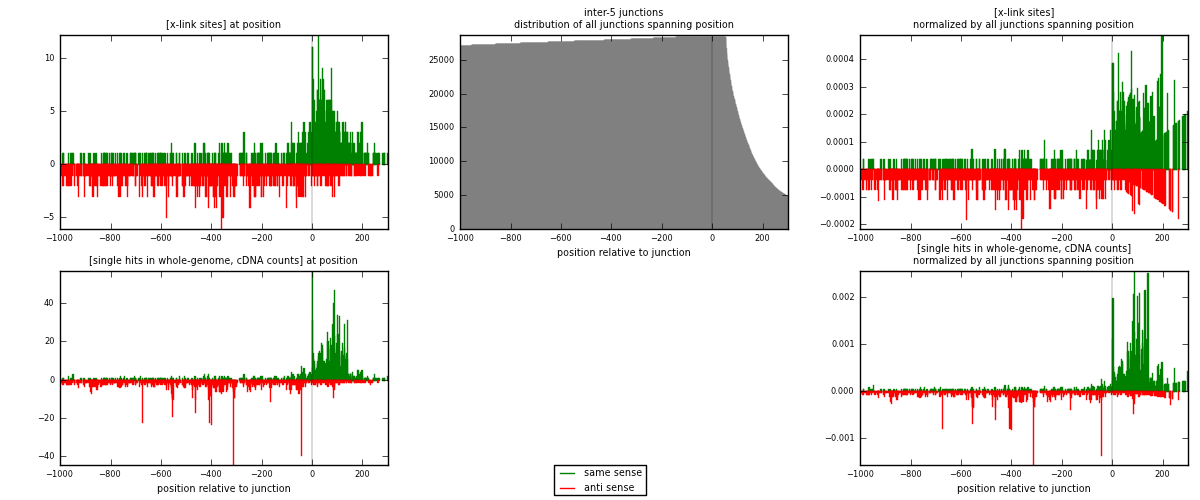
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 914 | 1.04% | 5.24% | 49.03% | 459 | 0.931702 |
| anti | 950 | 1.08% | 5.45% | 50.97% | 527 | 0.9684 |
| both | 1864 | 2.12% | 10.69% | 100.00% | 981 | 1.9001 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1976 | 2.00% | 9.29% | 57.93% | 459 | 2.01427 |
| anti | 1435 | 1.45% | 6.74% | 42.07% | 527 | 1.46279 |
| both | 3411 | 3.45% | 16.03% | 100.00% | 981 | 3.47706 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
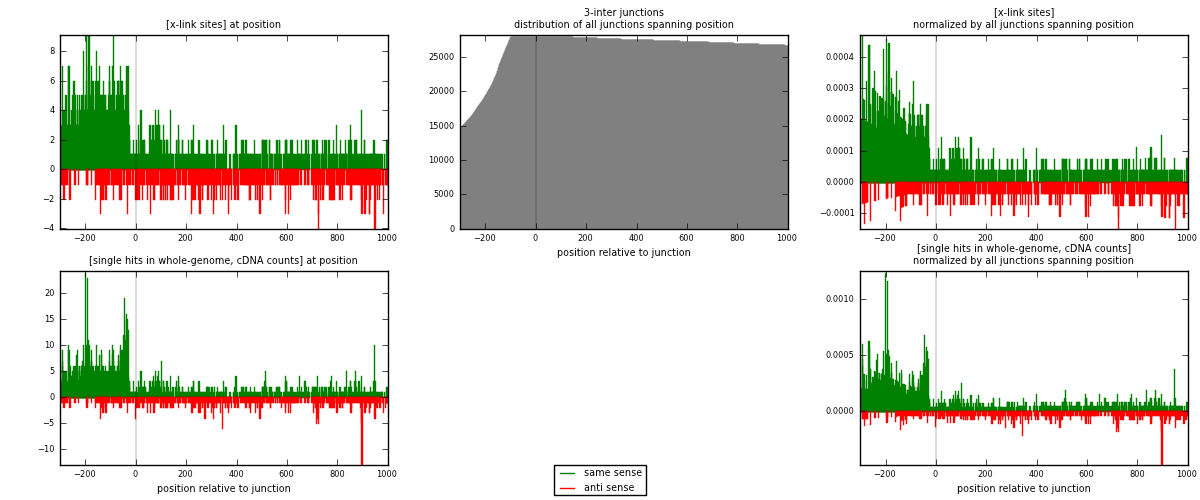
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1545 | 1.76% | 8.86% | 71.00% | 940 | 1.16078 |
| anti | 631 | 0.72% | 3.62% | 29.00% | 405 | 0.47408 |
| both | 2176 | 2.47% | 12.48% | 100.00% | 1331 | 1.63486 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1946 | 1.97% | 9.15% | 73.46% | 940 | 1.46206 |
| anti | 703 | 0.71% | 3.30% | 26.54% | 405 | 0.528174 |
| both | 2649 | 2.68% | 12.45% | 100.00% | 1331 | 1.99023 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
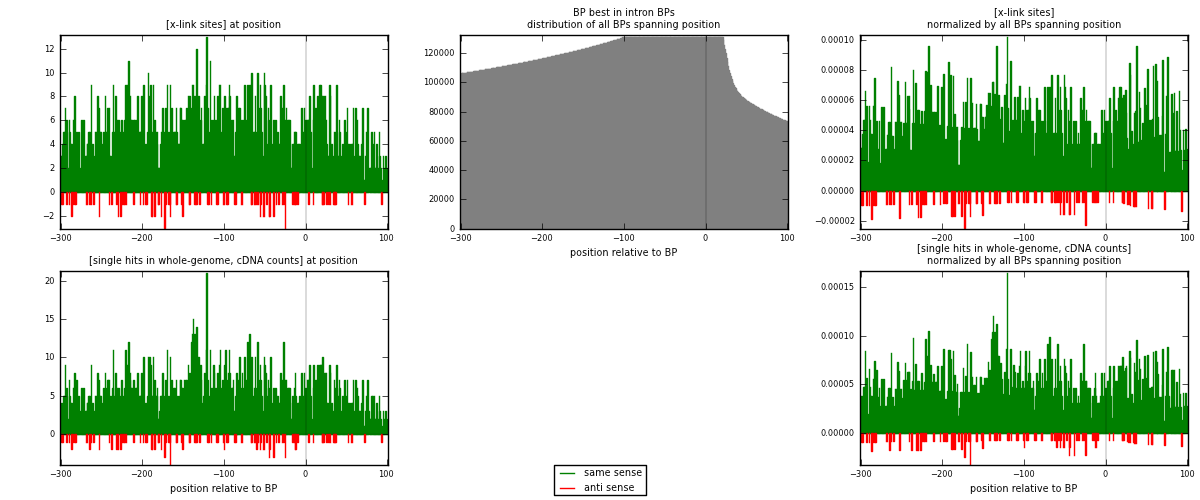
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 2002 | 2.28% | 11.48% | 94.21% | 1493 | 1.2458 |
| anti | 123 | 0.14% | 0.71% | 5.79% | 114 | 0.0765401 |
| both | 2125 | 2.42% | 12.18% | 100.00% | 1607 | 1.32234 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 2189 | 2.21% | 10.29% | 94.23% | 1493 | 1.36217 |
| anti | 134 | 0.14% | 0.63% | 5.77% | 114 | 0.0833852 |
| both | 2323 | 2.35% | 10.92% | 100.00% | 1607 | 1.44555 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.