RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
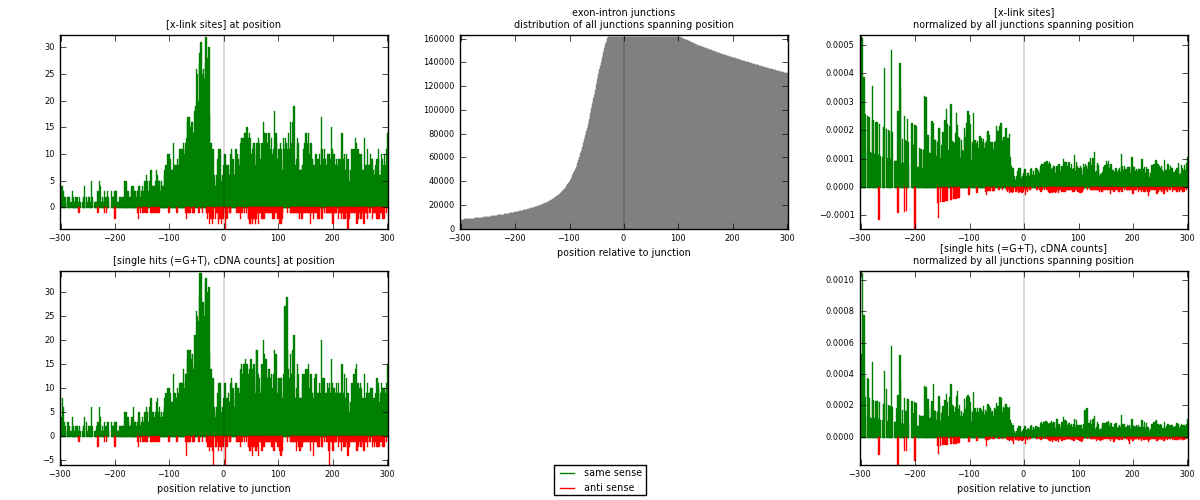 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4340 | 4.49% | 23.29% | 93.29% | 3380 | 1.1989 |
| anti | 312 | 0.32% | 1.67% | 6.71% | 244 | 0.0861878 |
| both | 4652 | 4.81% | 24.96% | 100.00% | 3620 | 1.28508 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4772 | 4.26% | 20.46% | 93.18% | 3380 | 1.31823 |
| anti | 349 | 0.31% | 1.50% | 6.82% | 244 | 0.0964088 |
| both | 5121 | 4.58% | 21.96% | 100.00% | 3620 | 1.41464 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
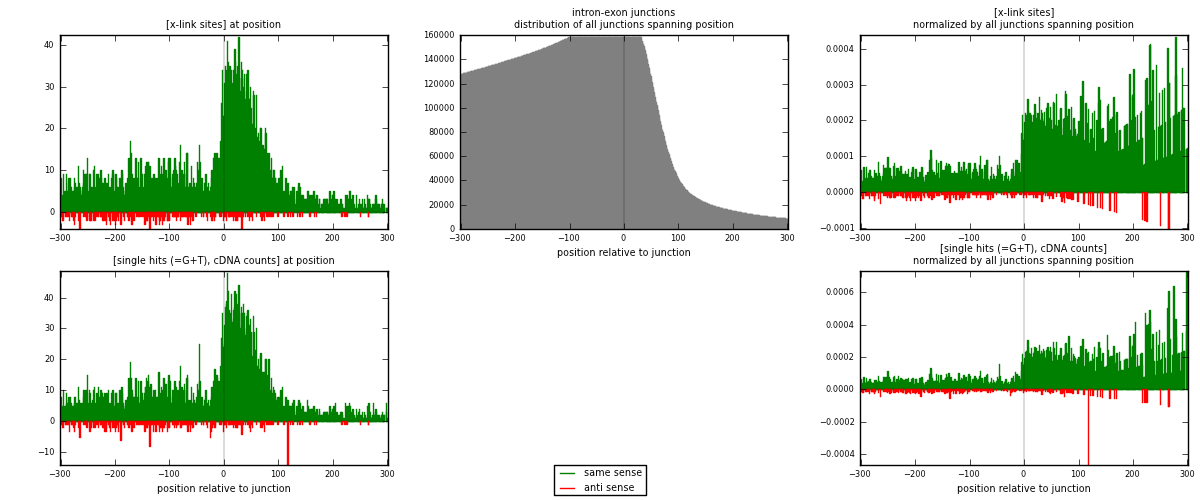 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4908 | 5.08% | 26.33% | 94.17% | 3947 | 1.17248 |
| anti | 304 | 0.31% | 1.63% | 5.83% | 248 | 0.072623 |
| both | 5212 | 5.39% | 27.96% | 100.00% | 4186 | 1.2451 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 5305 | 4.74% | 22.75% | 93.84% | 3947 | 1.26732 |
| anti | 348 | 0.31% | 1.49% | 6.16% | 248 | 0.0831343 |
| both | 5653 | 5.05% | 24.24% | 100.00% | 4186 | 1.35045 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
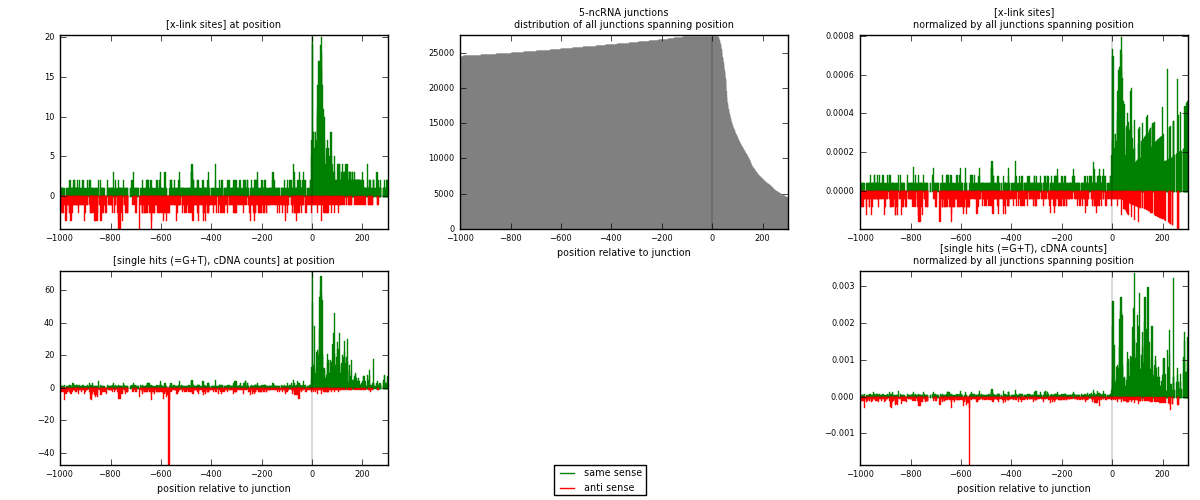 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1452 | 1.50% | 7.79% | 66.91% | 586 | 1.40154 |
| anti | 718 | 0.74% | 3.85% | 33.09% | 460 | 0.69305 |
| both | 2170 | 2.25% | 11.64% | 100.00% | 1036 | 2.09459 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3184 | 2.84% | 13.65% | 78.39% | 586 | 3.07336 |
| anti | 878 | 0.78% | 3.76% | 21.61% | 460 | 0.84749 |
| both | 4062 | 3.63% | 17.42% | 100.00% | 1036 | 3.92085 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
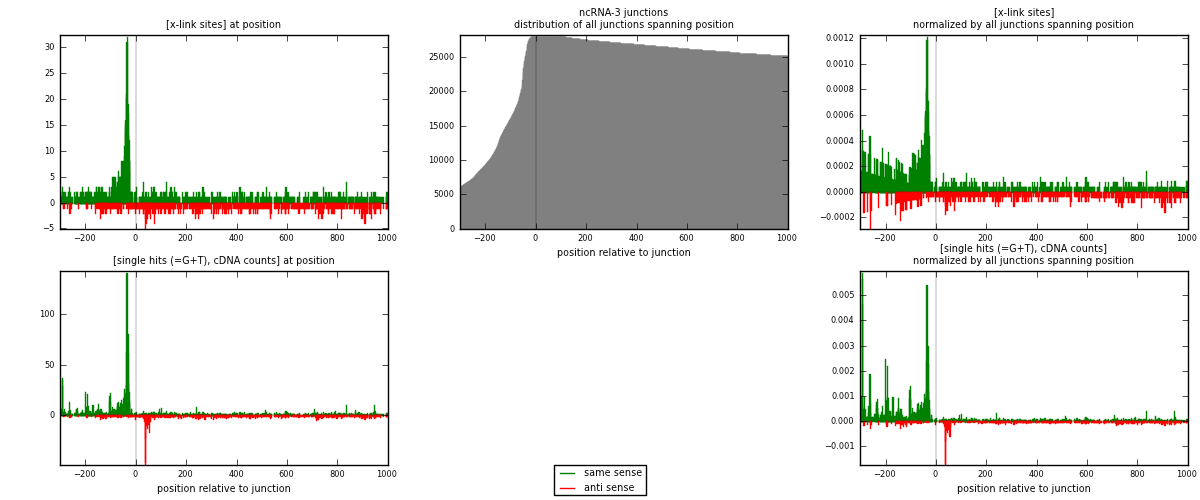 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1267 | 1.31% | 6.80% | 66.97% | 526 | 1.45465 |
| anti | 625 | 0.65% | 3.35% | 33.03% | 351 | 0.717566 |
| both | 1892 | 1.96% | 10.15% | 100.00% | 871 | 2.17222 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2316 | 2.07% | 9.93% | 72.72% | 526 | 2.65901 |
| anti | 869 | 0.78% | 3.73% | 27.28% | 351 | 0.997704 |
| both | 3185 | 2.85% | 13.66% | 100.00% | 871 | 3.65672 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
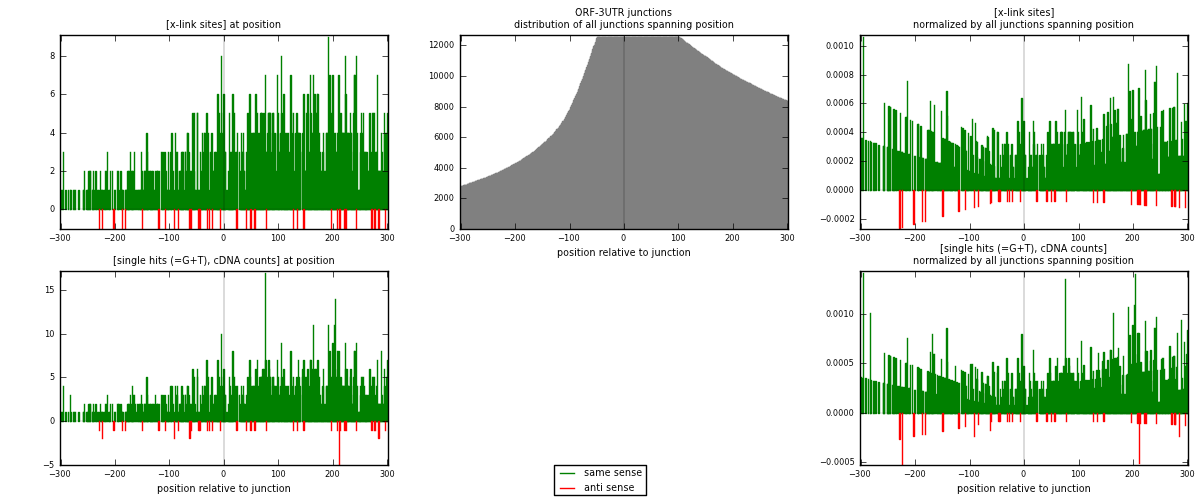
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1343 | 1.39% | 7.21% | 97.25% | 863 | 1.50392 |
| anti | 38 | 0.04% | 0.20% | 2.75% | 32 | 0.0425532 |
| both | 1381 | 1.43% | 7.41% | 100.00% | 893 | 1.54647 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1514 | 1.35% | 6.49% | 97.05% | 863 | 1.69541 |
| anti | 46 | 0.04% | 0.20% | 2.95% | 32 | 0.0515118 |
| both | 1560 | 1.39% | 6.69% | 100.00% | 893 | 1.74692 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
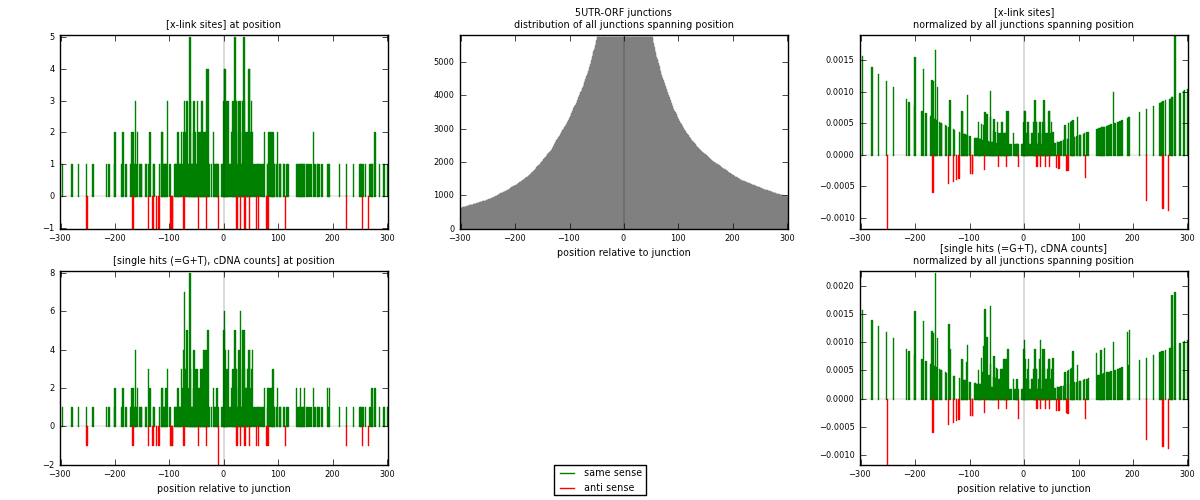
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 335 | 0.35% | 1.80% | 93.06% | 219 | 1.39583 |
| anti | 25 | 0.03% | 0.13% | 6.94% | 23 | 0.104167 |
| both | 360 | 0.37% | 1.93% | 100.00% | 240 | 1.5 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 395 | 0.35% | 1.69% | 93.82% | 219 | 1.64583 |
| anti | 26 | 0.02% | 0.11% | 6.18% | 23 | 0.108333 |
| both | 421 | 0.38% | 1.81% | 100.00% | 240 | 1.75417 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
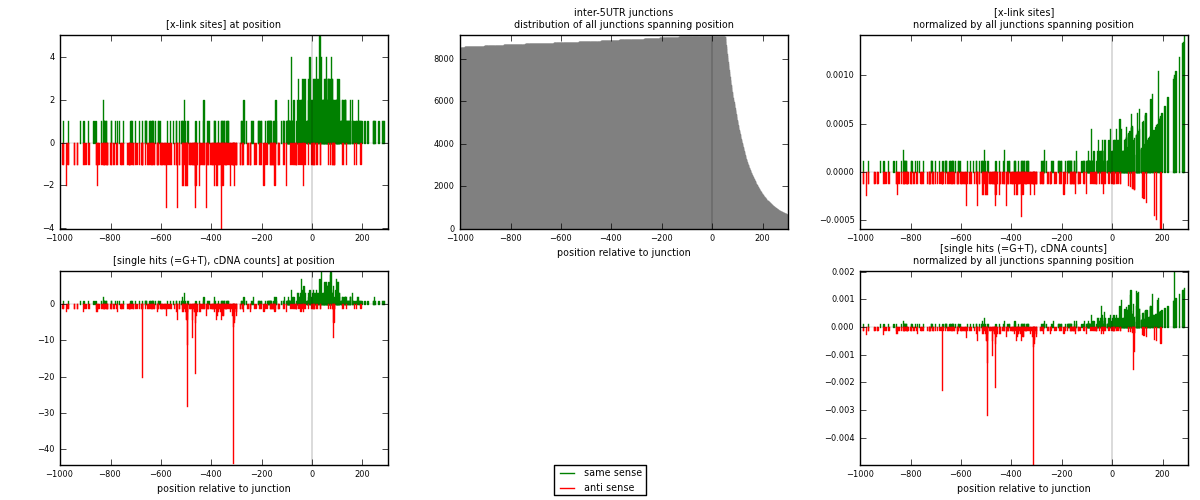
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 424 | 0.44% | 2.27% | 59.72% | 219 | 1.1013 |
| anti | 286 | 0.30% | 1.53% | 40.28% | 166 | 0.742857 |
| both | 710 | 0.73% | 3.81% | 100.00% | 385 | 1.84416 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 534 | 0.48% | 2.29% | 52.77% | 219 | 1.38701 |
| anti | 478 | 0.43% | 2.05% | 47.23% | 166 | 1.24156 |
| both | 1012 | 0.90% | 4.34% | 100.00% | 385 | 2.62857 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
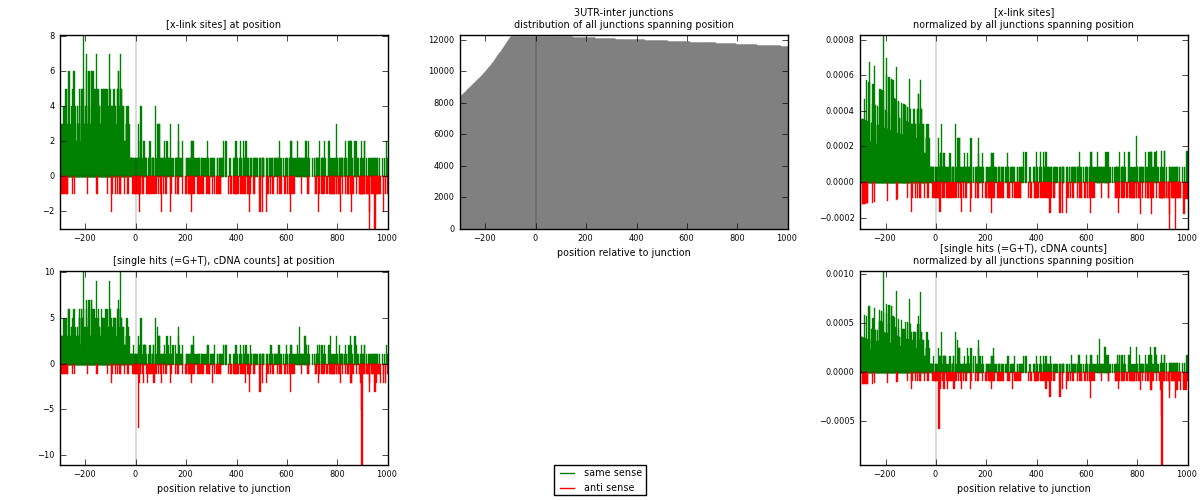
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1115 | 1.15% | 5.98% | 82.29% | 772 | 1.18997 |
| anti | 240 | 0.25% | 1.29% | 17.71% | 175 | 0.256137 |
| both | 1355 | 1.40% | 7.27% | 100.00% | 937 | 1.4461 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1225 | 1.09% | 5.25% | 81.67% | 772 | 1.30736 |
| anti | 275 | 0.25% | 1.18% | 18.33% | 175 | 0.29349 |
| both | 1500 | 1.34% | 6.43% | 100.00% | 937 | 1.60085 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
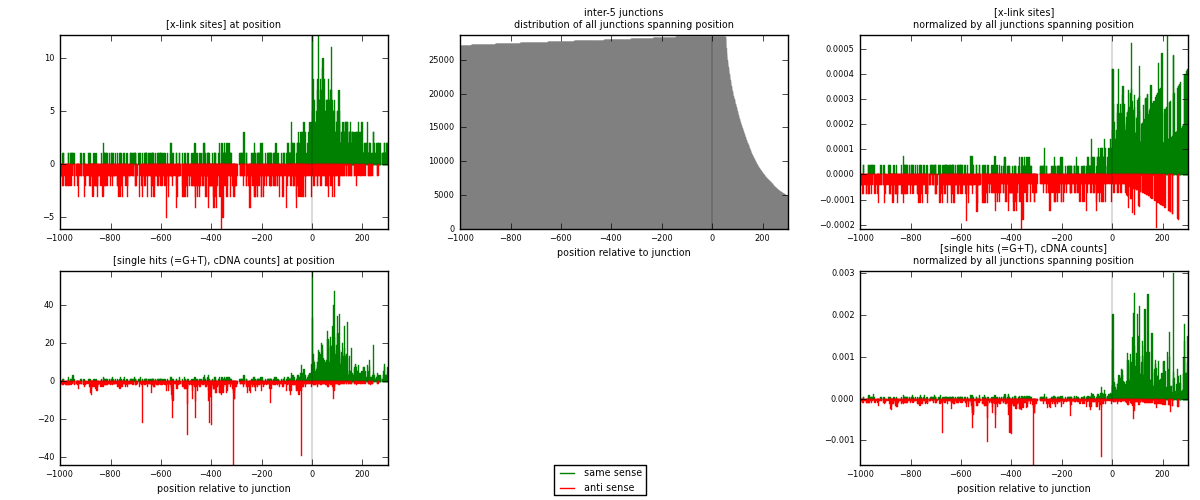
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1121 | 1.16% | 6.01% | 52.88% | 478 | 1.09259 |
| anti | 999 | 1.03% | 5.36% | 47.12% | 553 | 0.973684 |
| both | 2120 | 2.19% | 11.37% | 100.00% | 1026 | 2.06628 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2391 | 2.14% | 10.25% | 60.96% | 478 | 2.33041 |
| anti | 1531 | 1.37% | 6.56% | 39.04% | 553 | 1.4922 |
| both | 3922 | 3.50% | 16.82% | 100.00% | 1026 | 3.82261 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
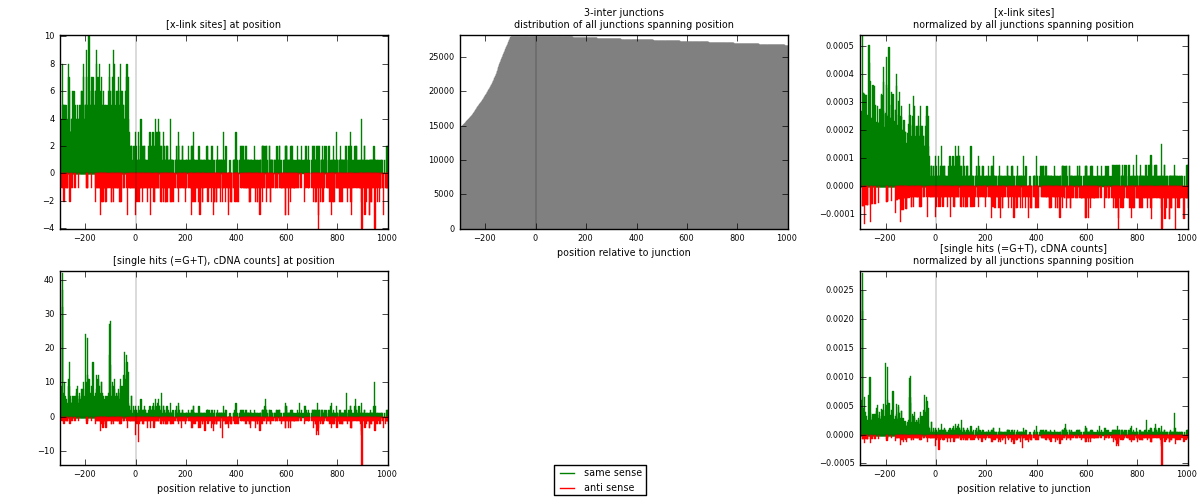
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1700 | 1.76% | 9.12% | 71.10% | 977 | 1.21602 |
| anti | 691 | 0.72% | 3.71% | 28.90% | 437 | 0.494278 |
| both | 2391 | 2.47% | 12.83% | 100.00% | 1398 | 1.7103 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2356 | 2.11% | 10.10% | 75.32% | 977 | 1.68526 |
| anti | 772 | 0.69% | 3.31% | 24.68% | 437 | 0.552217 |
| both | 3128 | 2.79% | 13.41% | 100.00% | 1398 | 2.23748 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
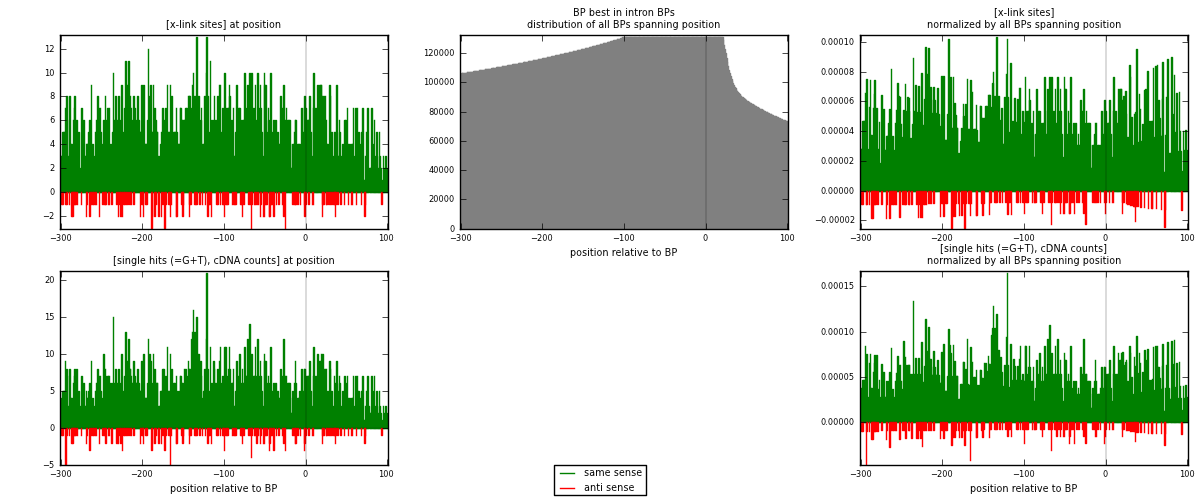
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 2128 | 2.20% | 11.42% | 91.45% | 1589 | 1.20498 |
| anti | 199 | 0.21% | 1.07% | 8.55% | 179 | 0.112684 |
| both | 2327 | 2.41% | 12.49% | 100.00% | 1766 | 1.31767 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 2326 | 2.08% | 9.97% | 91.36% | 1589 | 1.3171 |
| anti | 220 | 0.20% | 0.94% | 8.64% | 179 | 0.124575 |
| both | 2546 | 2.27% | 10.92% | 100.00% | 1766 | 1.44168 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.