RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
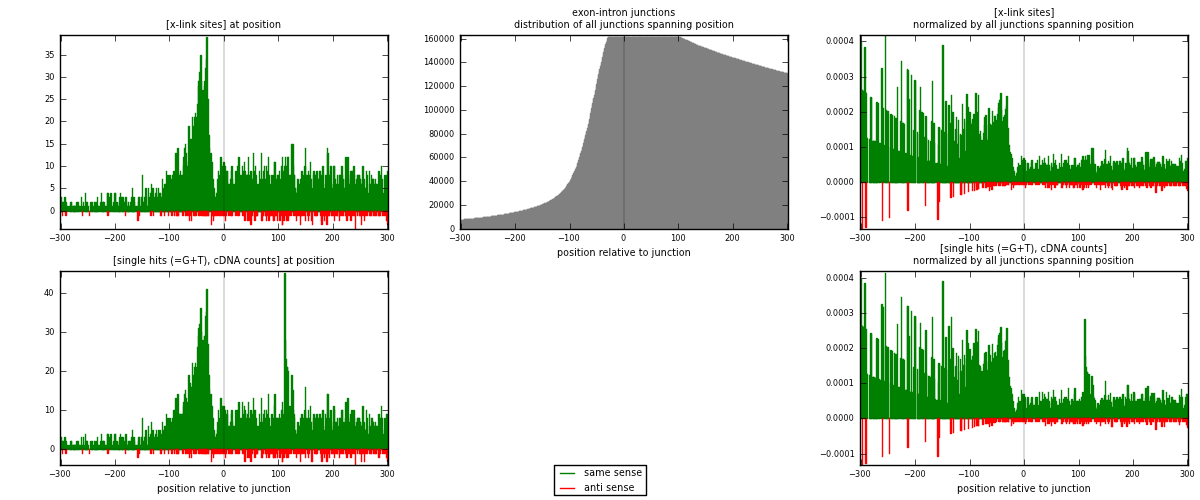 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3732 | 4.14% | 22.13% | 93.72% | 3439 | 1.01523 |
| anti | 250 | 0.28% | 1.48% | 6.28% | 240 | 0.0680087 |
| both | 3982 | 4.41% | 23.61% | 100.00% | 3676 | 1.08324 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3967 | 3.50% | 16.31% | 93.92% | 3439 | 1.07916 |
| anti | 257 | 0.23% | 1.06% | 6.08% | 240 | 0.0699129 |
| both | 4224 | 3.73% | 17.37% | 100.00% | 3676 | 1.14908 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
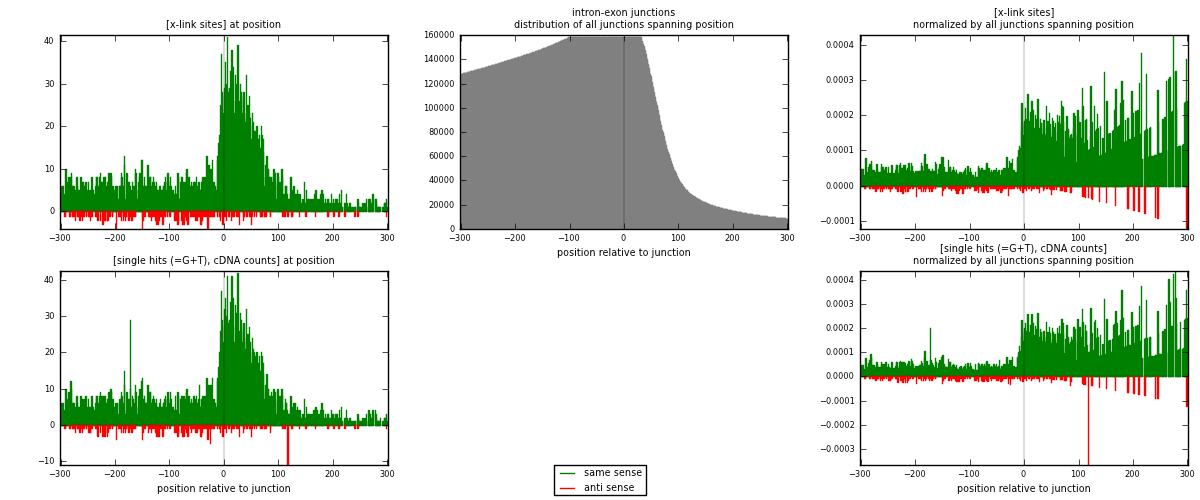 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4111 | 4.56% | 24.37% | 94.29% | 3825 | 1.01406 |
| anti | 249 | 0.28% | 1.48% | 5.71% | 236 | 0.0614208 |
| both | 4360 | 4.83% | 25.85% | 100.00% | 4054 | 1.07548 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4259 | 3.76% | 17.51% | 94.00% | 3825 | 1.05057 |
| anti | 272 | 0.24% | 1.12% | 6.00% | 236 | 0.0670942 |
| both | 4531 | 4.00% | 18.63% | 100.00% | 4054 | 1.11766 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
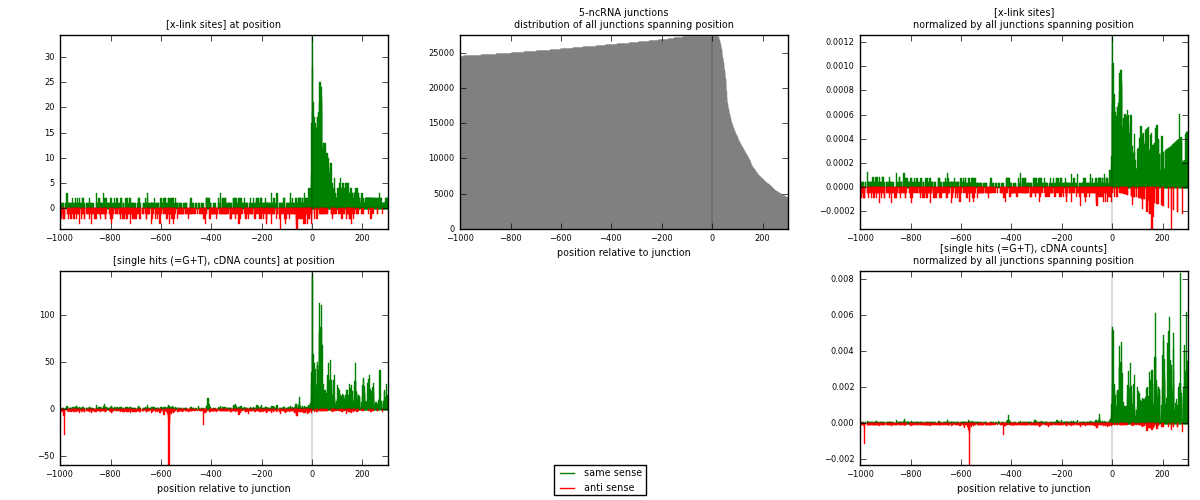 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2022 | 2.24% | 11.99% | 76.62% | 646 | 1.79255 |
| anti | 617 | 0.68% | 3.66% | 23.38% | 493 | 0.546986 |
| both | 2639 | 2.92% | 15.65% | 100.00% | 1128 | 2.33954 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 5691 | 5.02% | 23.40% | 87.76% | 646 | 5.04521 |
| anti | 794 | 0.70% | 3.26% | 12.24% | 493 | 0.703901 |
| both | 6485 | 5.72% | 26.66% | 100.00% | 1128 | 5.74911 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
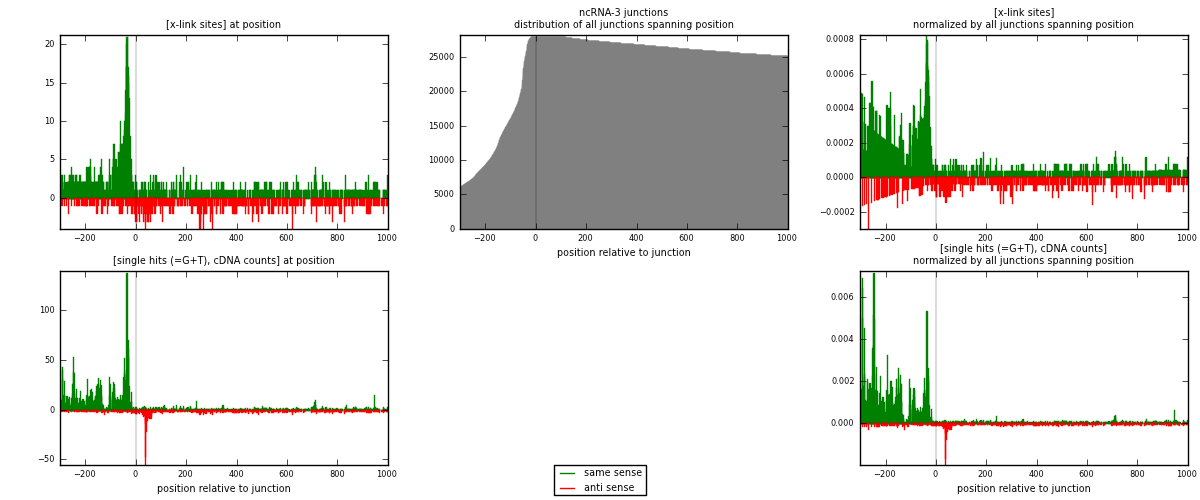 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1470 | 1.63% | 8.72% | 71.85% | 516 | 1.59783 |
| anti | 576 | 0.64% | 3.42% | 28.15% | 409 | 0.626087 |
| both | 2046 | 2.27% | 12.13% | 100.00% | 920 | 2.22391 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3881 | 3.43% | 15.96% | 82.59% | 516 | 4.21848 |
| anti | 818 | 0.72% | 3.36% | 17.41% | 409 | 0.88913 |
| both | 4699 | 4.15% | 19.32% | 100.00% | 920 | 5.10761 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
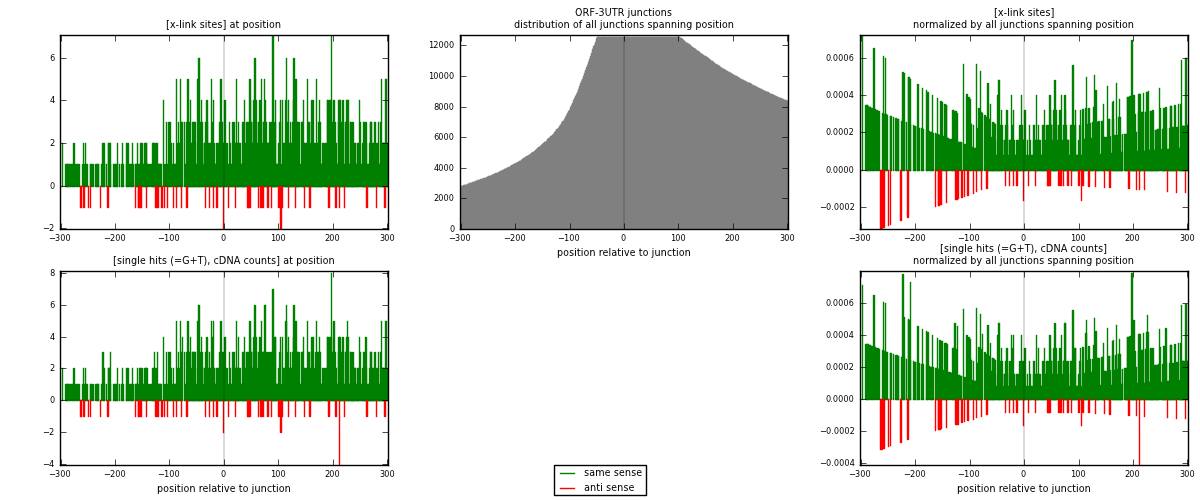
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 938 | 1.04% | 5.56% | 94.46% | 761 | 1.17104 |
| anti | 55 | 0.06% | 0.33% | 5.54% | 43 | 0.0686642 |
| both | 993 | 1.10% | 5.89% | 100.00% | 801 | 1.2397 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 967 | 0.85% | 3.98% | 94.34% | 761 | 1.20724 |
| anti | 58 | 0.05% | 0.24% | 5.66% | 43 | 0.0724095 |
| both | 1025 | 0.90% | 4.21% | 100.00% | 801 | 1.27965 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
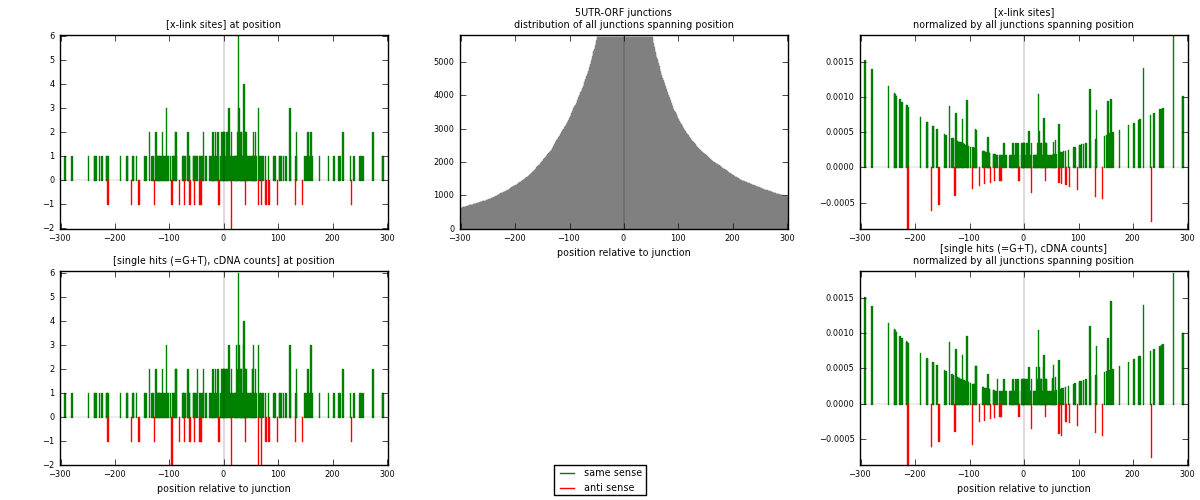
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 211 | 0.23% | 1.25% | 89.41% | 170 | 1.12234 |
| anti | 25 | 0.03% | 0.15% | 10.59% | 18 | 0.132979 |
| both | 236 | 0.26% | 1.40% | 100.00% | 188 | 1.25532 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 223 | 0.20% | 0.92% | 88.84% | 170 | 1.18617 |
| anti | 28 | 0.02% | 0.12% | 11.16% | 18 | 0.148936 |
| both | 251 | 0.22% | 1.03% | 100.00% | 188 | 1.33511 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
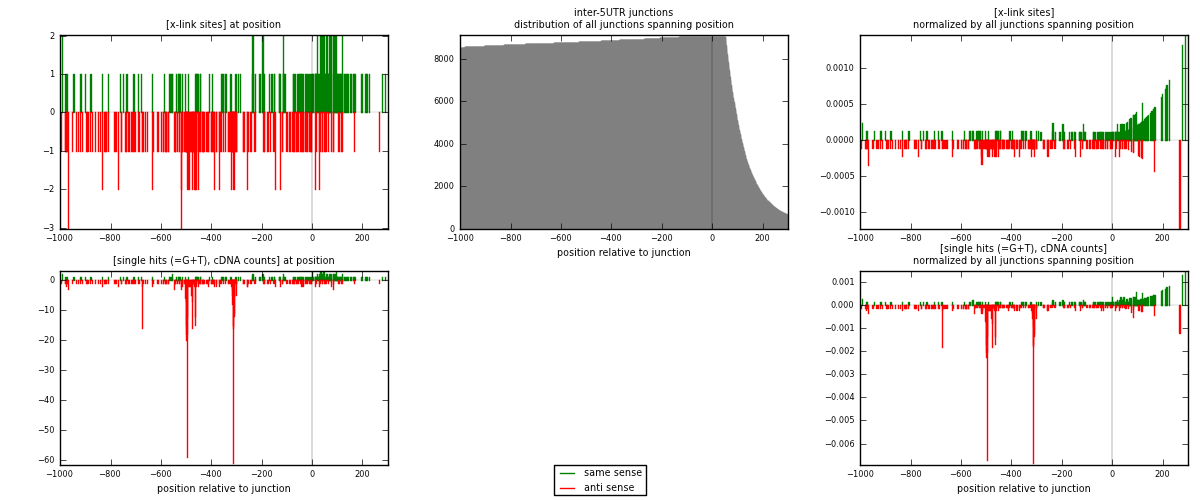
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 171 | 0.19% | 1.01% | 42.01% | 154 | 0.555195 |
| anti | 236 | 0.26% | 1.40% | 57.99% | 159 | 0.766234 |
| both | 407 | 0.45% | 2.41% | 100.00% | 308 | 1.32143 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 179 | 0.16% | 0.74% | 23.55% | 154 | 0.581169 |
| anti | 581 | 0.51% | 2.39% | 76.45% | 159 | 1.88636 |
| both | 760 | 0.67% | 3.12% | 100.00% | 308 | 2.46753 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 914 | 1.01% | 5.42% | 81.46% | 767 | 0.987041 |
| anti | 208 | 0.23% | 1.23% | 18.54% | 166 | 0.224622 |
| both | 1122 | 1.24% | 6.65% | 100.00% | 926 | 1.21166 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 938 | 0.83% | 3.86% | 81.00% | 767 | 1.01296 |
| anti | 220 | 0.19% | 0.90% | 19.00% | 166 | 0.237581 |
| both | 1158 | 1.02% | 4.76% | 100.00% | 926 | 1.25054 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
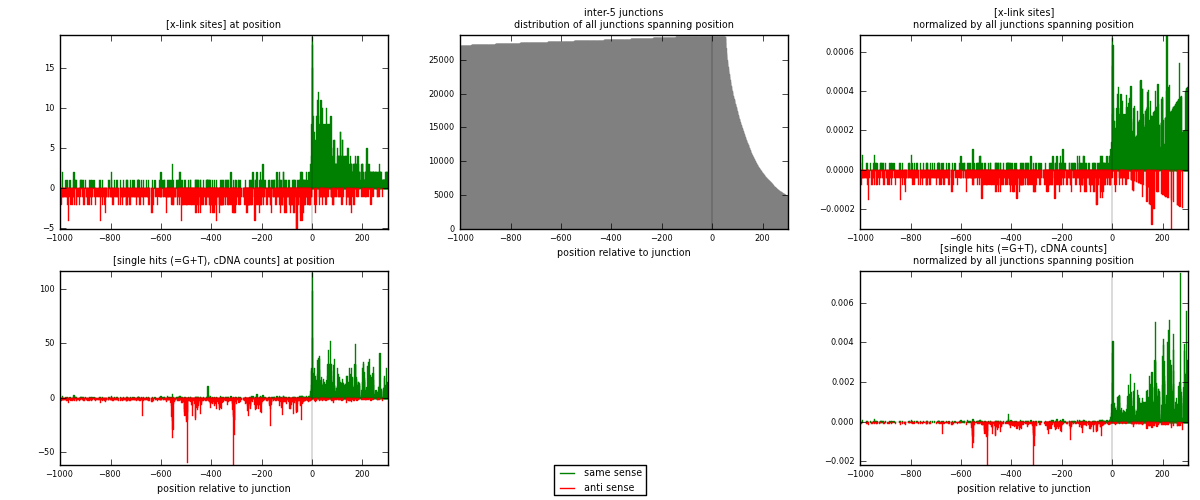
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1223 | 1.36% | 7.25% | 56.36% | 437 | 1.23911 |
| anti | 947 | 1.05% | 5.61% | 43.64% | 560 | 0.959473 |
| both | 2170 | 2.40% | 12.87% | 100.00% | 987 | 2.19858 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4045 | 3.57% | 16.63% | 66.29% | 437 | 4.09828 |
| anti | 2057 | 1.82% | 8.46% | 33.71% | 560 | 2.08409 |
| both | 6102 | 5.39% | 25.09% | 100.00% | 987 | 6.18237 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
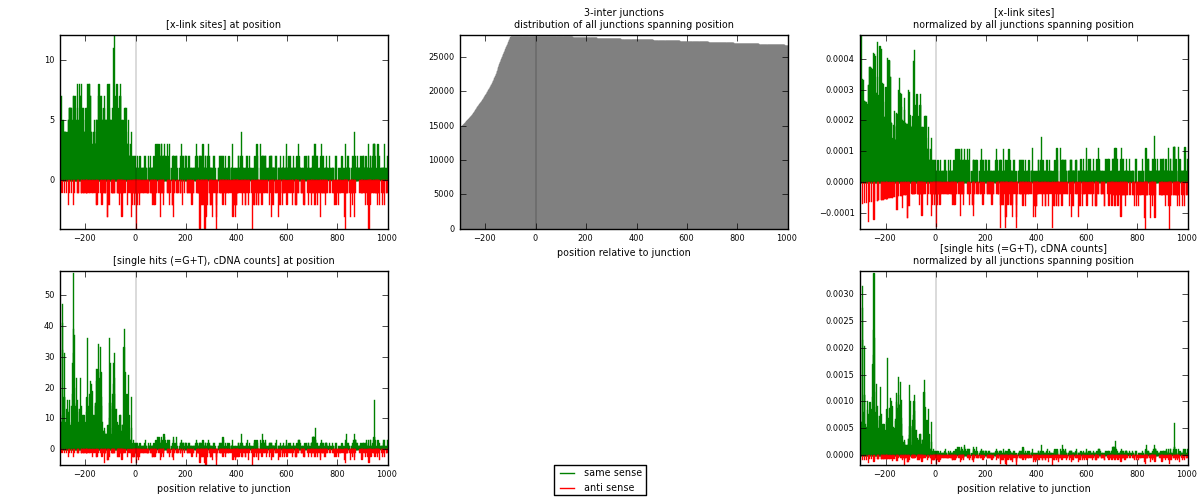 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1803 | 2.00% | 10.69% | 75.09% | 979 | 1.27331 |
| anti | 598 | 0.66% | 3.55% | 24.91% | 445 | 0.422316 |
| both | 2401 | 2.66% | 14.24% | 100.00% | 1416 | 1.69562 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3784 | 3.34% | 15.56% | 85.65% | 979 | 2.67232 |
| anti | 634 | 0.56% | 2.61% | 14.35% | 445 | 0.44774 |
| both | 4418 | 3.90% | 18.16% | 100.00% | 1416 | 3.12006 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
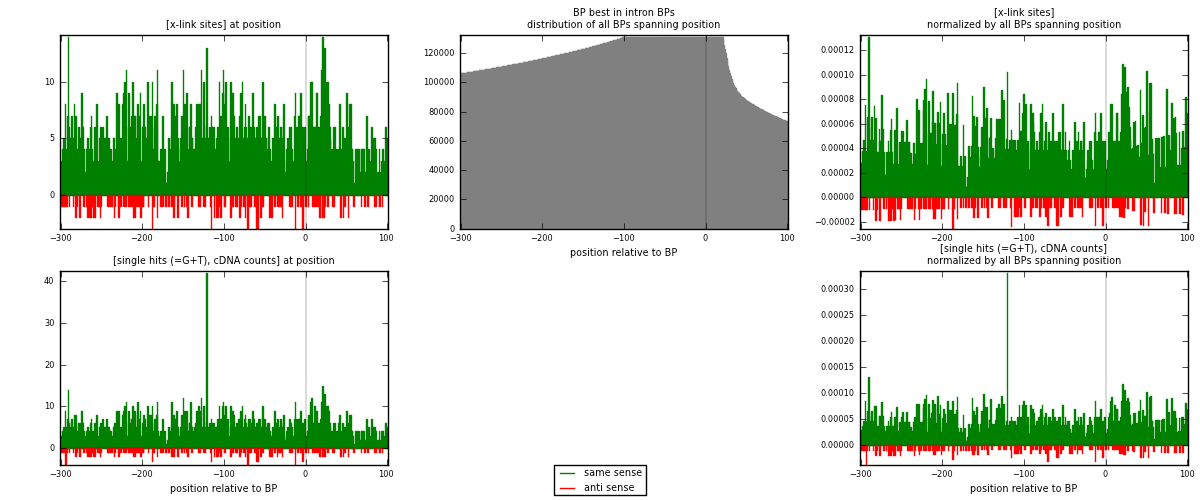 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 2074 | 2.30% | 12.30% | 91.37% | 1805 | 1.03908 |
| anti | 196 | 0.22% | 1.16% | 8.63% | 192 | 0.0981964 |
| both | 2270 | 2.52% | 13.46% | 100.00% | 1996 | 1.13727 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 2186 | 1.93% | 8.99% | 91.46% | 1805 | 1.09519 |
| anti | 204 | 0.18% | 0.84% | 8.54% | 192 | 0.102204 |
| both | 2390 | 2.11% | 9.83% | 100.00% | 1996 | 1.19739 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.