RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
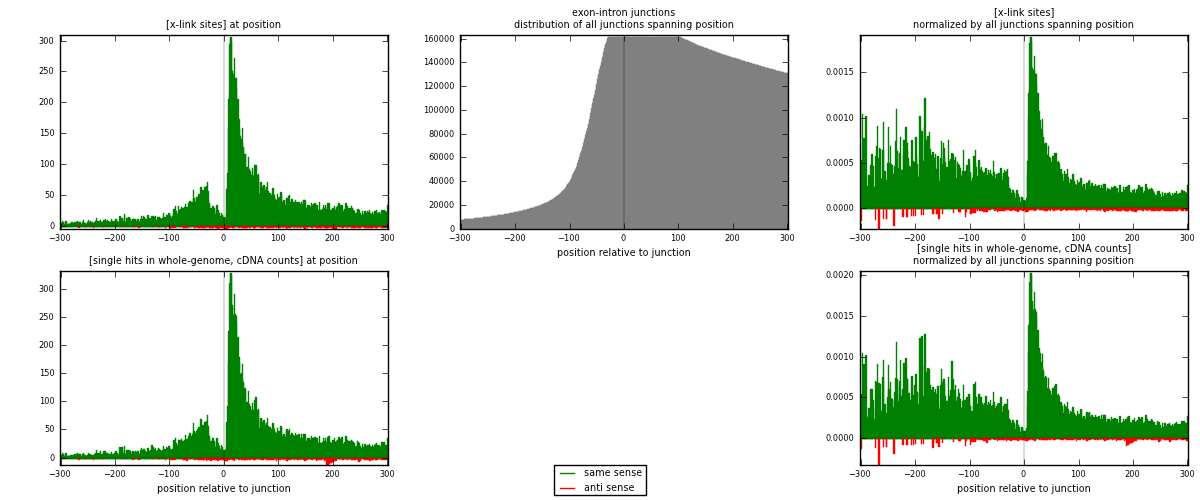 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 20754 | 9.96% | 30.78% | 97.63% | 15559 | 1.30528 |
| anti | 504 | 0.24% | 0.75% | 2.37% | 366 | 0.0316981 |
| both | 21258 | 10.21% | 31.53% | 100.00% | 15900 | 1.33698 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 21757 | 9.54% | 28.52% | 97.23% | 15559 | 1.36836 |
| anti | 620 | 0.27% | 0.81% | 2.77% | 366 | 0.0389937 |
| both | 22377 | 9.82% | 29.33% | 100.00% | 15900 | 1.40736 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
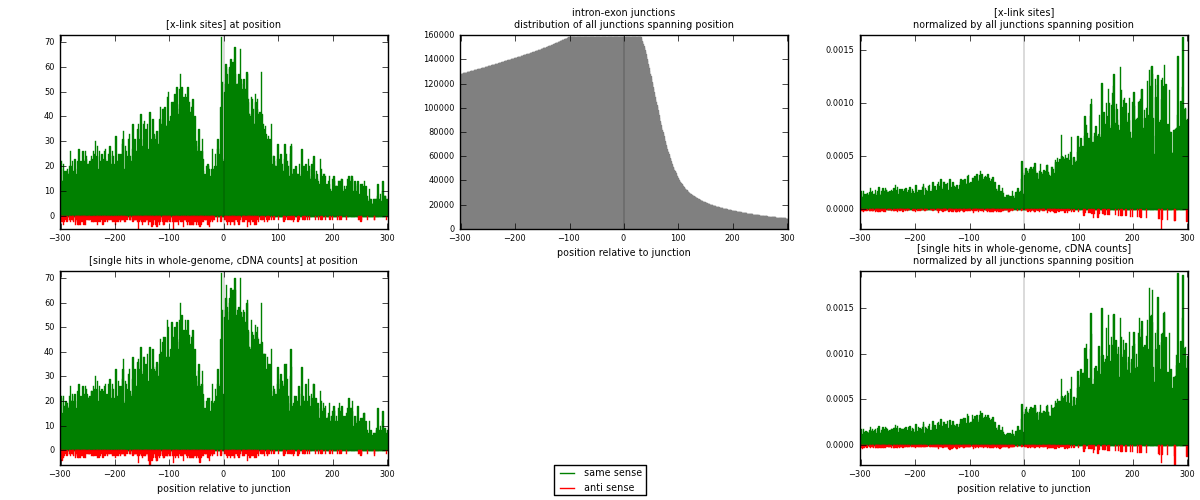 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 14822 | 7.12% | 21.98% | 96.74% | 11215 | 1.27864 |
| anti | 500 | 0.24% | 0.74% | 3.26% | 405 | 0.0431332 |
| both | 15322 | 7.36% | 22.73% | 100.00% | 11592 | 1.32177 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 15714 | 6.89% | 20.60% | 96.82% | 11215 | 1.35559 |
| anti | 516 | 0.23% | 0.68% | 3.18% | 405 | 0.0445135 |
| both | 16230 | 7.12% | 21.27% | 100.00% | 11592 | 1.4001 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
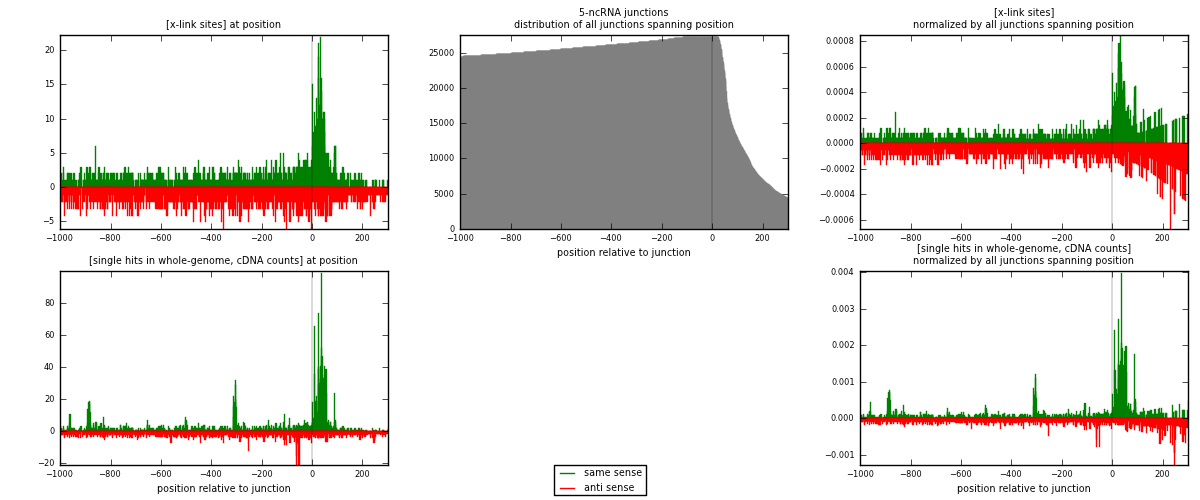 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1579 | 0.76% | 2.34% | 46.90% | 781 | 0.931563 |
| anti | 1788 | 0.86% | 2.65% | 53.10% | 940 | 1.05487 |
| both | 3367 | 1.62% | 4.99% | 100.00% | 1695 | 1.98643 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2931 | 1.29% | 3.84% | 59.10% | 781 | 1.7292 |
| anti | 2028 | 0.89% | 2.66% | 40.90% | 940 | 1.19646 |
| both | 4959 | 2.18% | 6.50% | 100.00% | 1695 | 2.92566 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
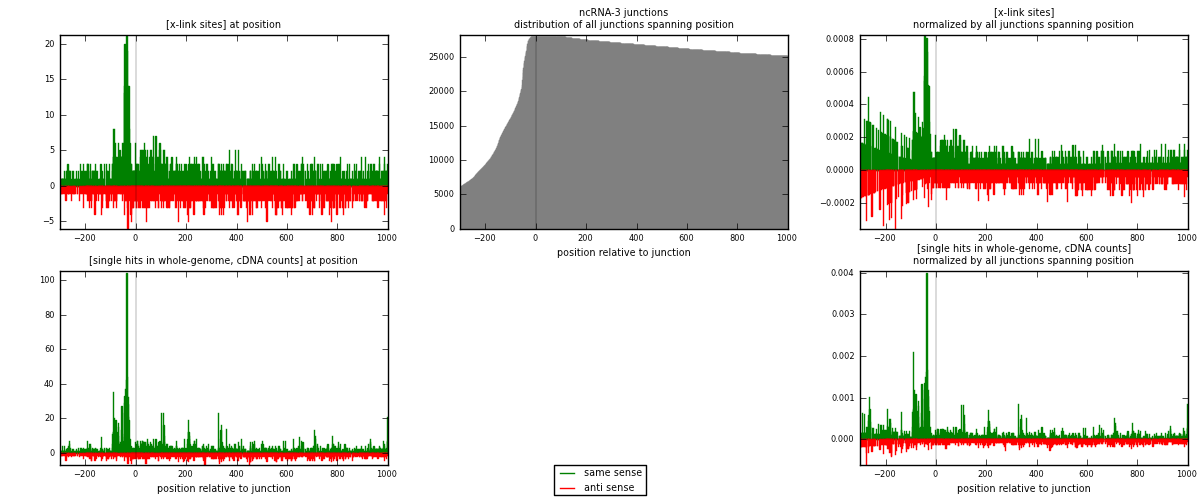 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1973 | 0.95% | 2.93% | 57.06% | 936 | 1.14178 |
| anti | 1485 | 0.71% | 2.20% | 42.94% | 807 | 0.859375 |
| both | 3458 | 1.66% | 5.13% | 100.00% | 1728 | 2.00116 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3165 | 1.39% | 4.15% | 66.53% | 936 | 1.8316 |
| anti | 1592 | 0.70% | 2.09% | 33.47% | 807 | 0.921296 |
| both | 4757 | 2.09% | 6.24% | 100.00% | 1728 | 2.75289 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
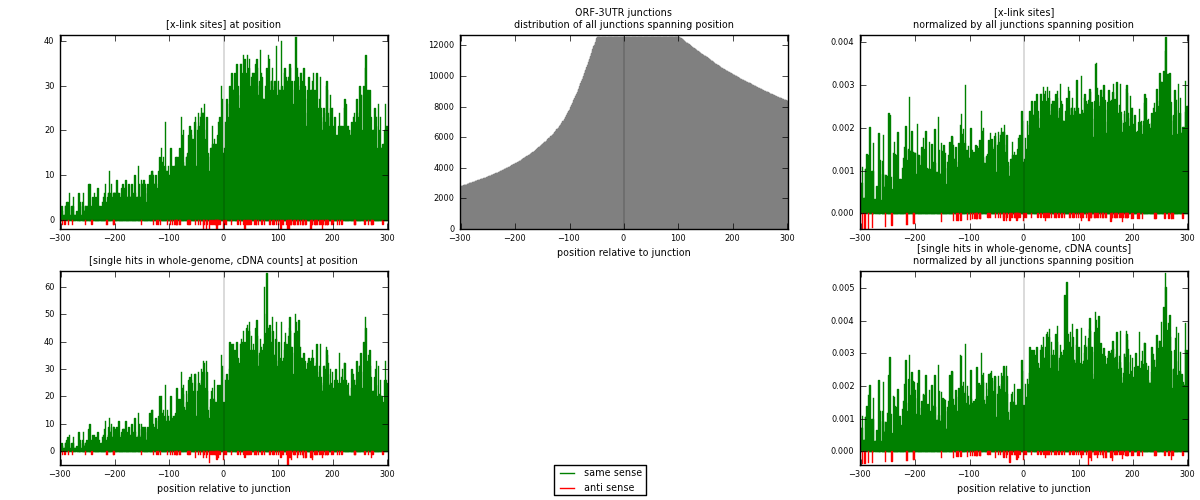
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10139 | 4.87% | 15.04% | 98.68% | 3180 | 3.12836 |
| anti | 136 | 0.07% | 0.20% | 1.32% | 85 | 0.0419624 |
| both | 10275 | 4.93% | 15.24% | 100.00% | 3241 | 3.17032 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 12210 | 5.36% | 16.00% | 98.74% | 3180 | 3.76736 |
| anti | 156 | 0.07% | 0.20% | 1.26% | 85 | 0.0481333 |
| both | 12366 | 5.42% | 16.21% | 100.00% | 3241 | 3.81549 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
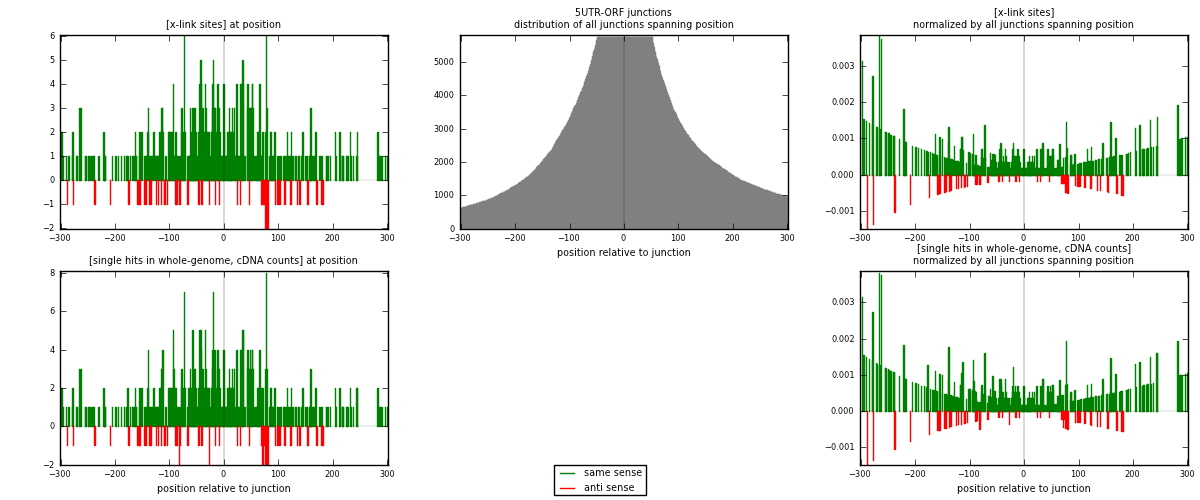
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 464 | 0.22% | 0.69% | 89.75% | 318 | 1.34493 |
| anti | 53 | 0.03% | 0.08% | 10.25% | 28 | 0.153623 |
| both | 517 | 0.25% | 0.77% | 100.00% | 345 | 1.49855 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 490 | 0.21% | 0.64% | 89.74% | 318 | 1.42029 |
| anti | 56 | 0.02% | 0.07% | 10.26% | 28 | 0.162319 |
| both | 546 | 0.24% | 0.72% | 100.00% | 345 | 1.58261 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
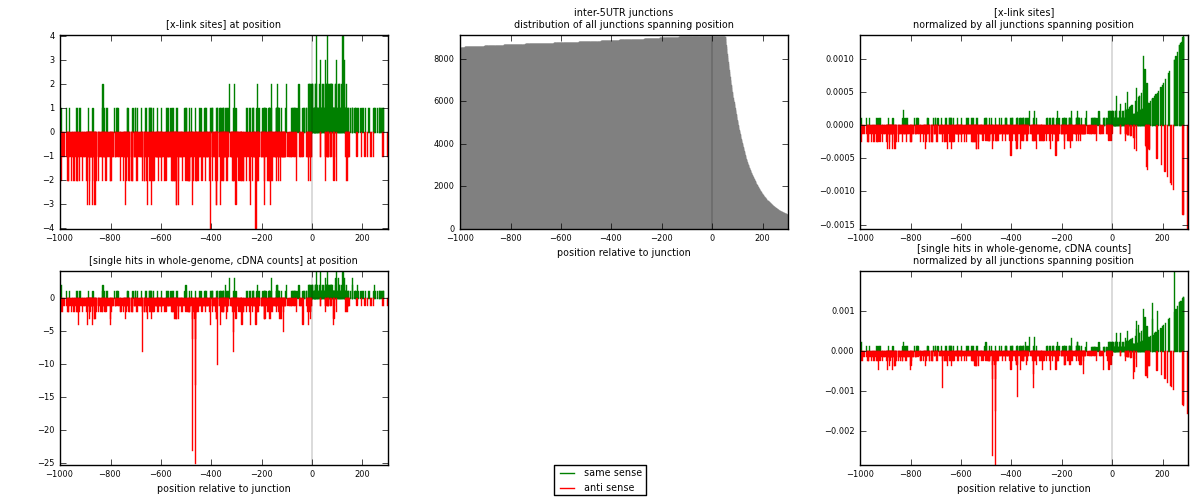
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 316 | 0.15% | 0.47% | 32.28% | 243 | 0.466765 |
| anti | 663 | 0.32% | 0.98% | 67.72% | 447 | 0.979321 |
| both | 979 | 0.47% | 1.45% | 100.00% | 677 | 1.44609 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 335 | 0.15% | 0.44% | 29.21% | 243 | 0.49483 |
| anti | 812 | 0.36% | 1.06% | 70.79% | 447 | 1.19941 |
| both | 1147 | 0.50% | 1.50% | 100.00% | 677 | 1.69424 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
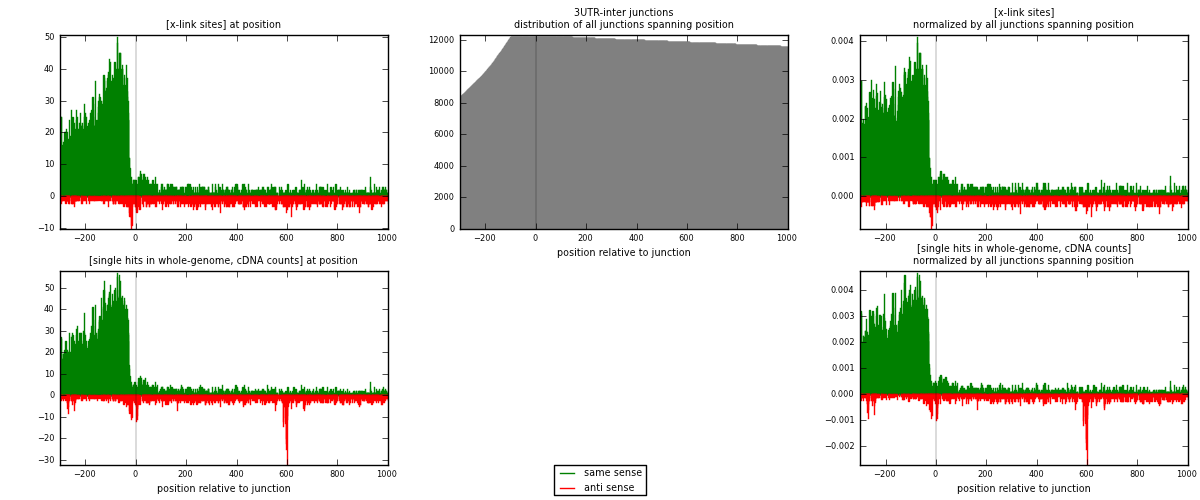
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8111 | 3.89% | 12.03% | 84.91% | 2946 | 2.4104 |
| anti | 1441 | 0.69% | 2.14% | 15.09% | 563 | 0.428232 |
| both | 9552 | 4.59% | 14.17% | 100.00% | 3365 | 2.83863 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9270 | 4.07% | 12.15% | 83.48% | 2946 | 2.75483 |
| anti | 1835 | 0.80% | 2.41% | 16.52% | 563 | 0.545319 |
| both | 11105 | 4.87% | 14.56% | 100.00% | 3365 | 3.30015 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
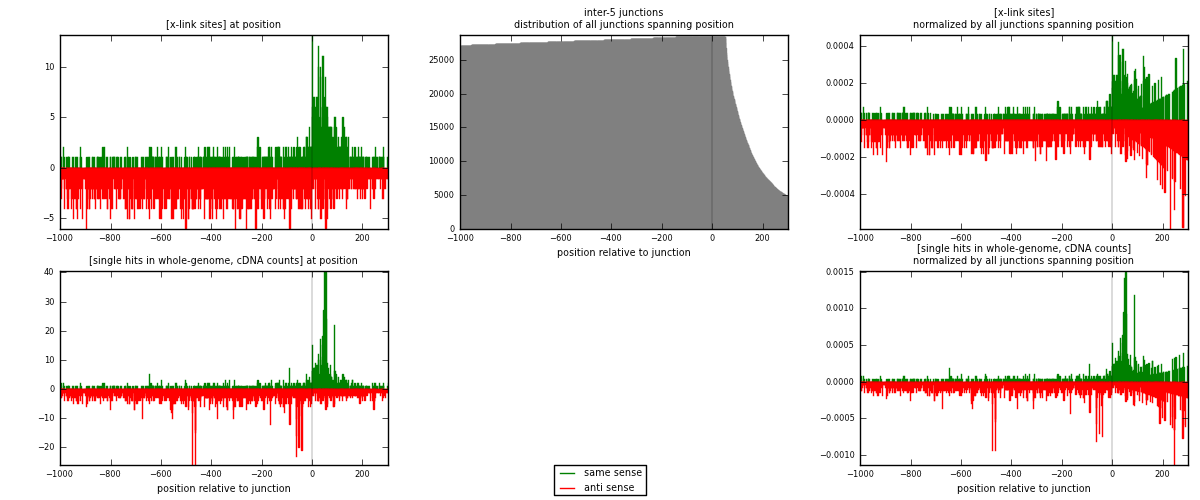
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 893 | 0.43% | 1.32% | 28.13% | 582 | 0.499441 |
| anti | 2281 | 1.10% | 3.38% | 71.87% | 1244 | 1.27573 |
| both | 3174 | 1.52% | 4.71% | 100.00% | 1788 | 1.77517 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1190 | 0.52% | 1.56% | 30.20% | 582 | 0.665548 |
| anti | 2750 | 1.21% | 3.60% | 69.80% | 1244 | 1.53803 |
| both | 3940 | 1.73% | 5.16% | 100.00% | 1788 | 2.20358 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
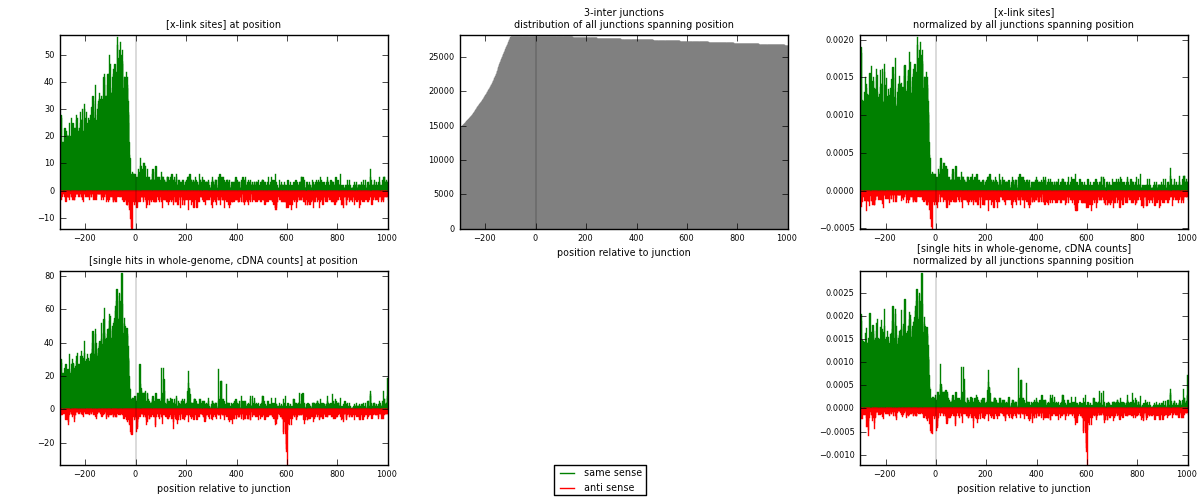
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9897 | 4.75% | 14.68% | 78.55% | 3595 | 2.17182 |
| anti | 2702 | 1.30% | 4.01% | 21.45% | 1133 | 0.592934 |
| both | 12599 | 6.05% | 18.69% | 100.00% | 4557 | 2.76476 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 12119 | 5.32% | 15.89% | 78.81% | 3595 | 2.65943 |
| anti | 3259 | 1.43% | 4.27% | 21.19% | 1133 | 0.715163 |
| both | 15378 | 6.75% | 20.16% | 100.00% | 4557 | 3.37459 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
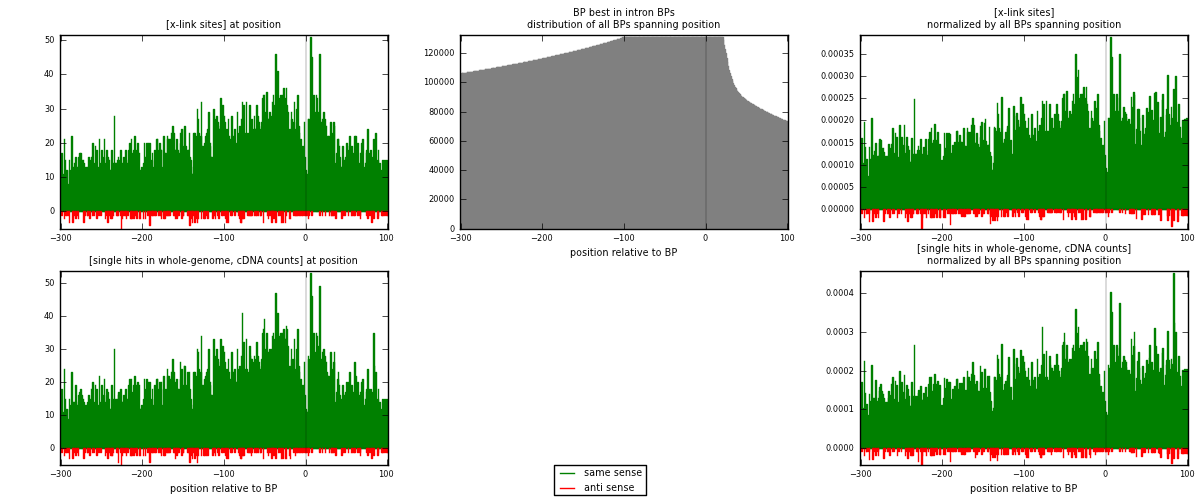
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 7989 | 3.84% | 11.85% | 96.40% | 6674 | 1.15582 |
| anti | 298 | 0.14% | 0.44% | 3.60% | 245 | 0.0431134 |
| both | 8287 | 3.98% | 12.29% | 100.00% | 6912 | 1.19893 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 8271 | 3.63% | 10.84% | 96.39% | 6674 | 1.19661 |
| anti | 310 | 0.14% | 0.41% | 3.61% | 245 | 0.0448495 |
| both | 8581 | 3.76% | 11.25% | 100.00% | 6912 | 1.24146 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.