RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
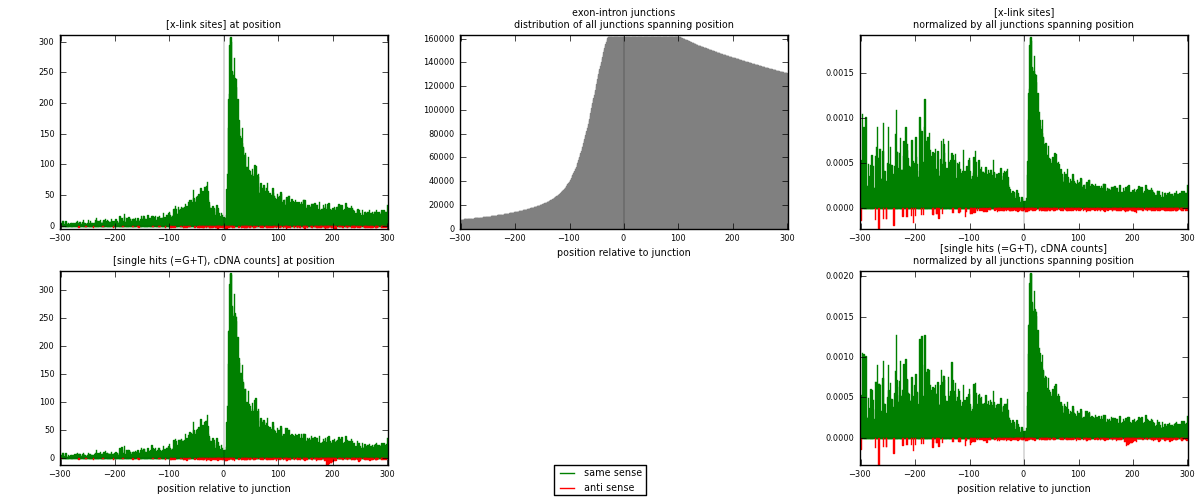 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 20942 | 9.74% | 30.28% | 97.58% | 15661 | 1.30806 |
| anti | 519 | 0.24% | 0.75% | 2.42% | 376 | 0.0324172 |
| both | 21461 | 9.98% | 31.03% | 100.00% | 16010 | 1.34047 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 21997 | 9.25% | 27.82% | 97.19% | 15661 | 1.37395 |
| anti | 636 | 0.27% | 0.80% | 2.81% | 376 | 0.0397252 |
| both | 22633 | 9.52% | 28.62% | 100.00% | 16010 | 1.41368 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
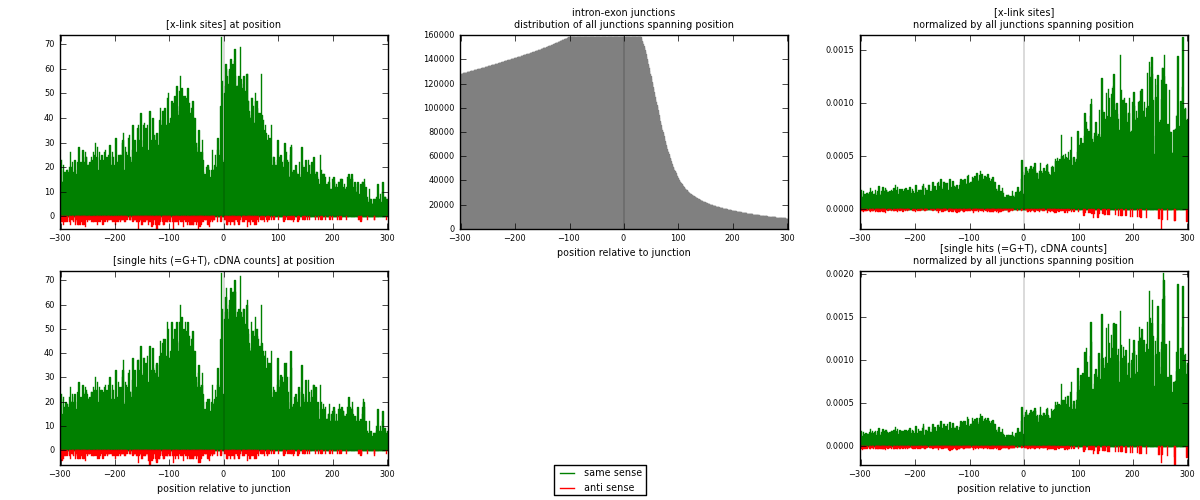 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 15005 | 6.98% | 21.69% | 96.64% | 11289 | 1.28412 |
| anti | 522 | 0.24% | 0.75% | 3.36% | 427 | 0.0446727 |
| both | 15527 | 7.22% | 22.45% | 100.00% | 11685 | 1.3288 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 15942 | 6.70% | 20.16% | 96.74% | 11289 | 1.36431 |
| anti | 538 | 0.23% | 0.68% | 3.26% | 427 | 0.0460419 |
| both | 16480 | 6.93% | 20.84% | 100.00% | 11685 | 1.41036 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
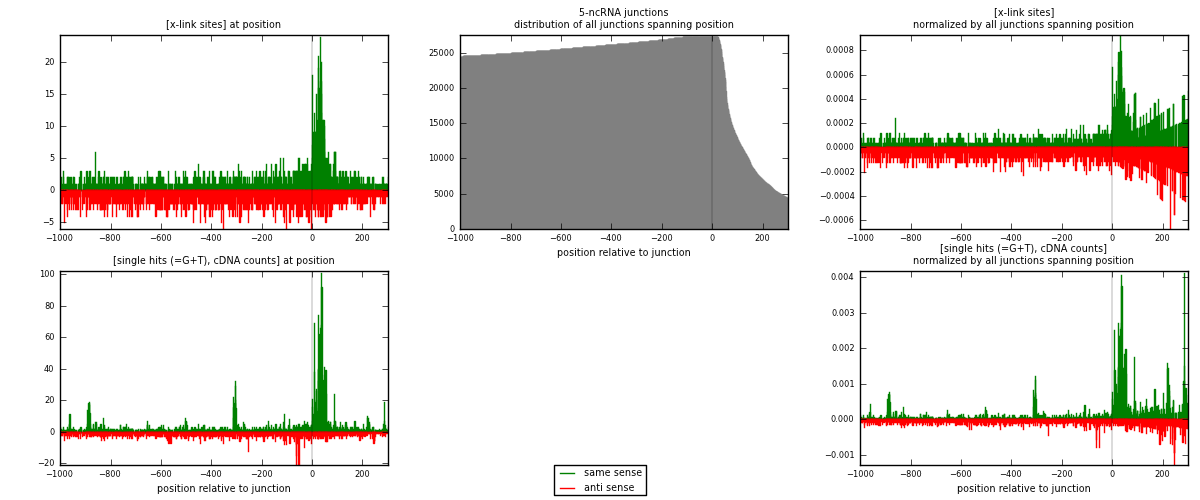 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1770 | 0.82% | 2.56% | 48.87% | 827 | 1.00625 |
| anti | 1852 | 0.86% | 2.68% | 51.13% | 961 | 1.05287 |
| both | 3622 | 1.68% | 5.24% | 100.00% | 1759 | 2.05912 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3468 | 1.46% | 4.39% | 62.13% | 827 | 1.97157 |
| anti | 2114 | 0.89% | 2.67% | 37.87% | 961 | 1.20182 |
| both | 5582 | 2.35% | 7.06% | 100.00% | 1759 | 3.17339 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
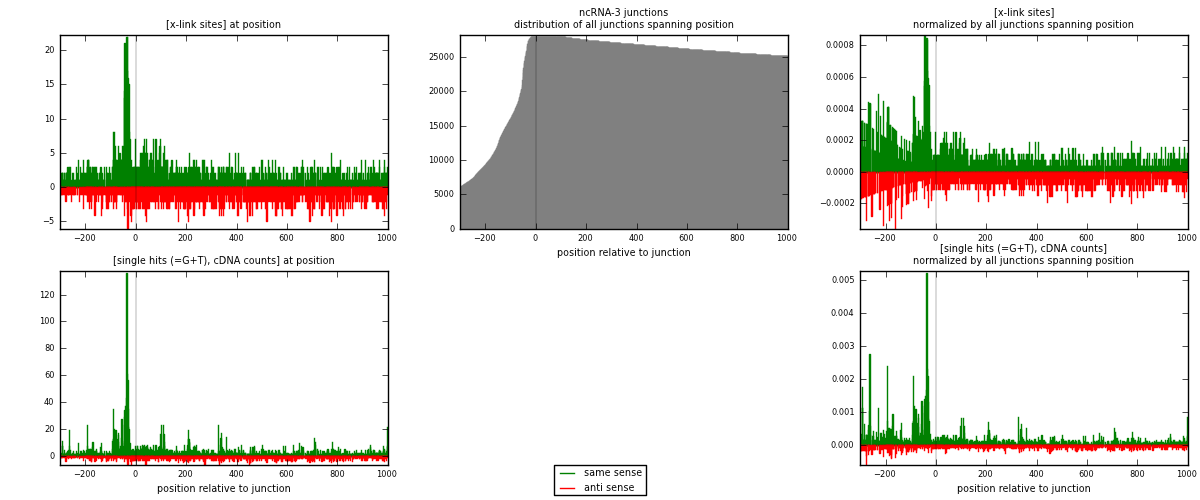 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2148 | 1.00% | 3.11% | 58.54% | 989 | 1.19799 |
| anti | 1521 | 0.71% | 2.20% | 41.46% | 821 | 0.848299 |
| both | 3669 | 1.71% | 5.30% | 100.00% | 1793 | 2.04629 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3536 | 1.49% | 4.47% | 68.46% | 989 | 1.97211 |
| anti | 1629 | 0.69% | 2.06% | 31.54% | 821 | 0.908533 |
| both | 5165 | 2.17% | 6.53% | 100.00% | 1793 | 2.88065 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
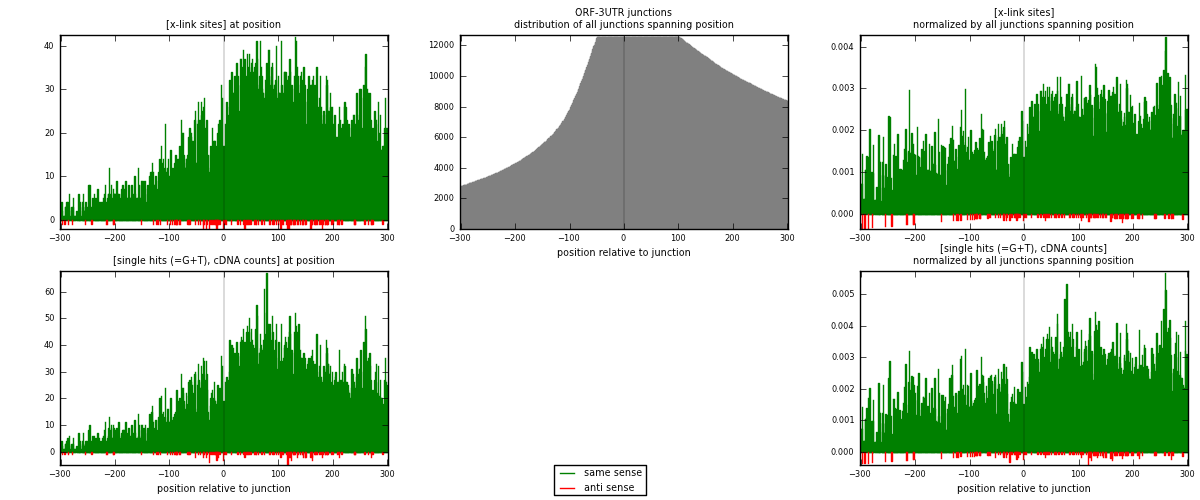
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10532 | 4.90% | 15.23% | 98.71% | 3219 | 3.21 |
| anti | 138 | 0.06% | 0.20% | 1.29% | 86 | 0.0420603 |
| both | 10670 | 4.96% | 15.43% | 100.00% | 3281 | 3.25206 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 12695 | 5.34% | 16.06% | 98.76% | 3219 | 3.86925 |
| anti | 159 | 0.07% | 0.20% | 1.24% | 86 | 0.0484608 |
| both | 12854 | 5.41% | 16.26% | 100.00% | 3281 | 3.91771 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
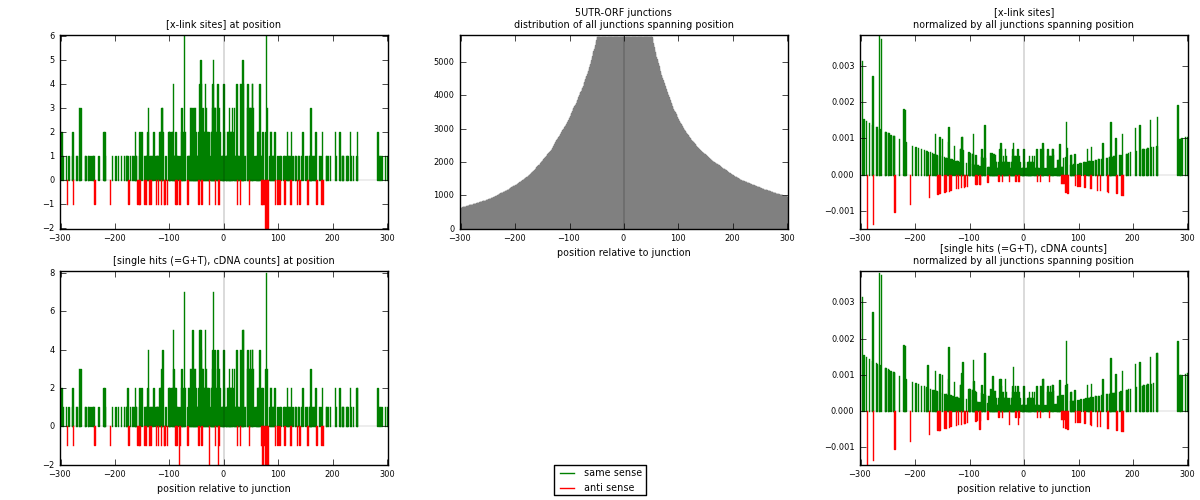
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 467 | 0.22% | 0.68% | 89.64% | 321 | 1.33811 |
| anti | 54 | 0.03% | 0.08% | 10.36% | 29 | 0.154728 |
| both | 521 | 0.24% | 0.75% | 100.00% | 349 | 1.49284 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 493 | 0.21% | 0.62% | 89.47% | 321 | 1.41261 |
| anti | 58 | 0.02% | 0.07% | 10.53% | 29 | 0.166189 |
| both | 551 | 0.23% | 0.70% | 100.00% | 349 | 1.5788 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
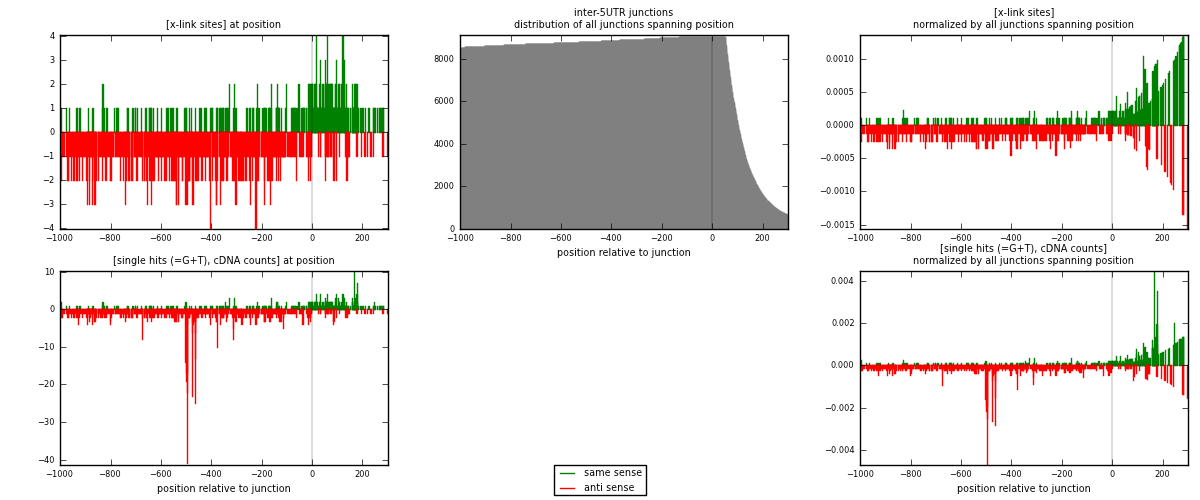
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 340 | 0.16% | 0.49% | 33.27% | 247 | 0.494186 |
| anti | 682 | 0.32% | 0.99% | 66.73% | 454 | 0.991279 |
| both | 1022 | 0.48% | 1.48% | 100.00% | 688 | 1.48547 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 379 | 0.16% | 0.48% | 28.73% | 247 | 0.550872 |
| anti | 940 | 0.40% | 1.19% | 71.27% | 454 | 1.36628 |
| both | 1319 | 0.55% | 1.67% | 100.00% | 688 | 1.91715 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
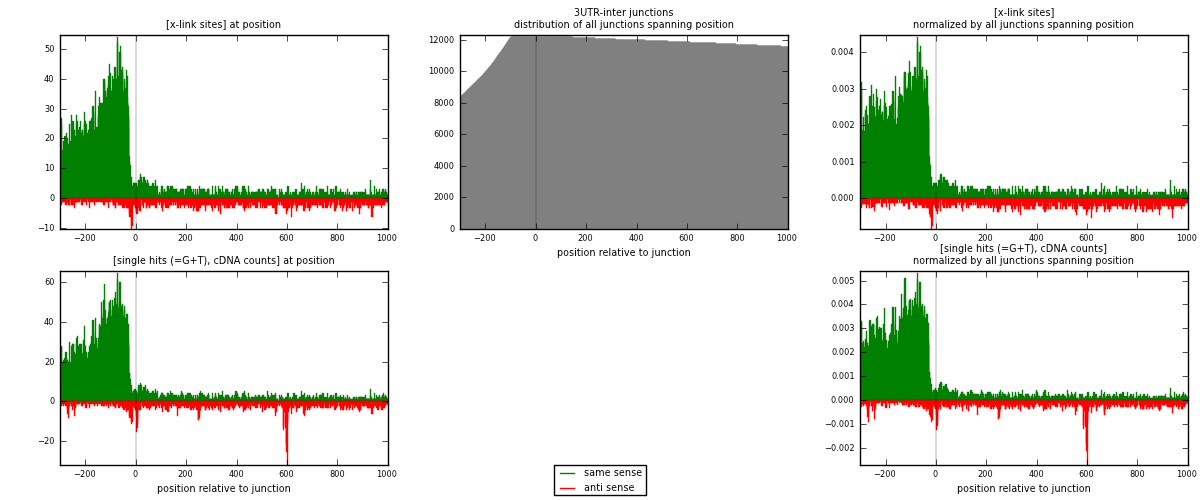
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8434 | 3.92% | 12.19% | 84.93% | 2994 | 2.4668 |
| anti | 1497 | 0.70% | 2.16% | 15.07% | 571 | 0.437847 |
| both | 9931 | 4.62% | 14.36% | 100.00% | 3419 | 2.90465 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 9691 | 4.08% | 12.26% | 83.32% | 2994 | 2.83445 |
| anti | 1940 | 0.82% | 2.45% | 16.68% | 571 | 0.567417 |
| both | 11631 | 4.89% | 14.71% | 100.00% | 3419 | 3.40187 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
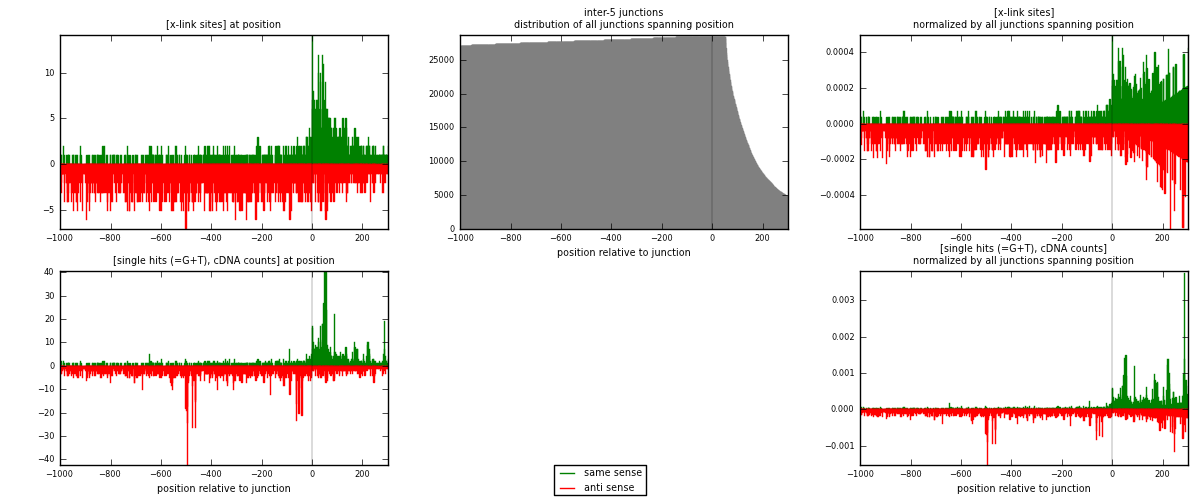
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1044 | 0.49% | 1.51% | 30.65% | 604 | 0.568627 |
| anti | 2362 | 1.10% | 3.42% | 69.35% | 1272 | 1.28649 |
| both | 3406 | 1.58% | 4.92% | 100.00% | 1836 | 1.85512 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1488 | 0.63% | 1.88% | 33.54% | 604 | 0.810458 |
| anti | 2949 | 1.24% | 3.73% | 66.46% | 1272 | 1.60621 |
| both | 4437 | 1.87% | 5.61% | 100.00% | 1836 | 2.41667 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
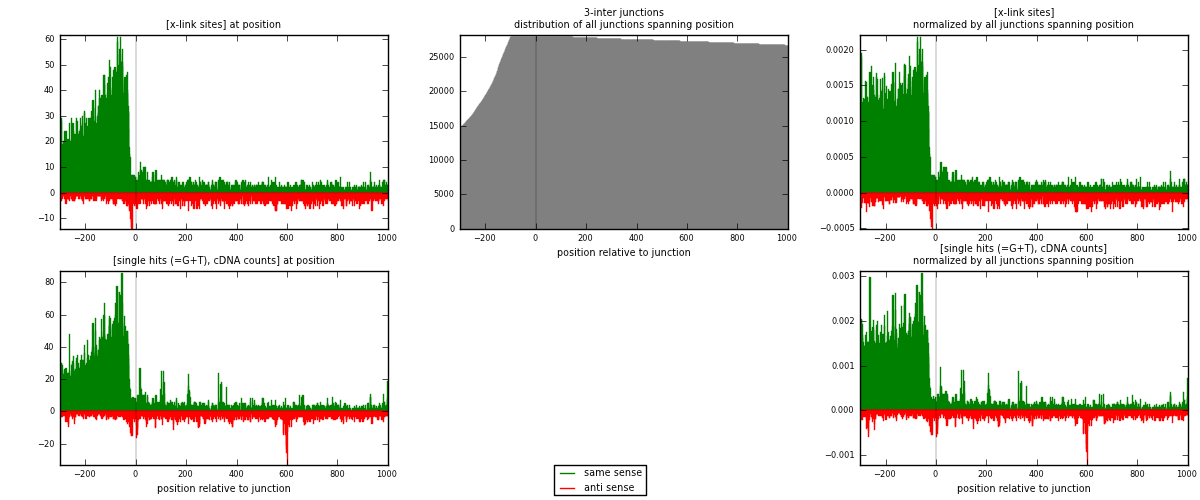
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10352 | 4.81% | 14.97% | 78.71% | 3674 | 2.22719 |
| anti | 2800 | 1.30% | 4.05% | 21.29% | 1147 | 0.60241 |
| both | 13152 | 6.12% | 19.02% | 100.00% | 4648 | 2.8296 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 12805 | 5.39% | 16.19% | 78.94% | 3674 | 2.75495 |
| anti | 3416 | 1.44% | 4.32% | 21.06% | 1147 | 0.73494 |
| both | 16221 | 6.82% | 20.51% | 100.00% | 4648 | 3.48989 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
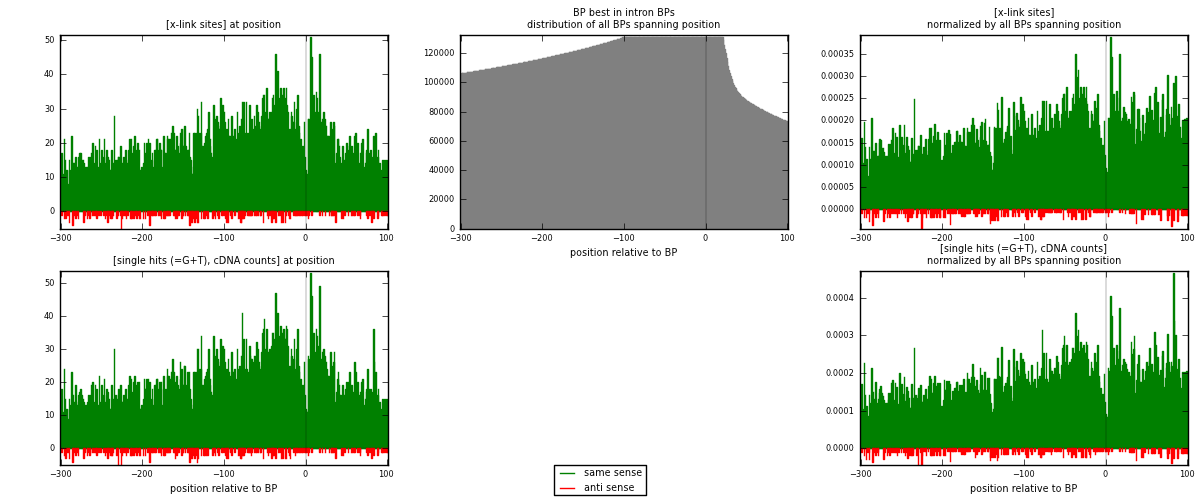
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 8050 | 3.74% | 11.64% | 96.18% | 6719 | 1.15346 |
| anti | 320 | 0.15% | 0.46% | 3.82% | 267 | 0.0458518 |
| both | 8370 | 3.89% | 12.10% | 100.00% | 6979 | 1.19931 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 8334 | 3.50% | 10.54% | 96.15% | 6719 | 1.19415 |
| anti | 334 | 0.14% | 0.42% | 3.85% | 267 | 0.0478579 |
| both | 8668 | 3.65% | 10.96% | 100.00% | 6979 | 1.24201 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.