RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
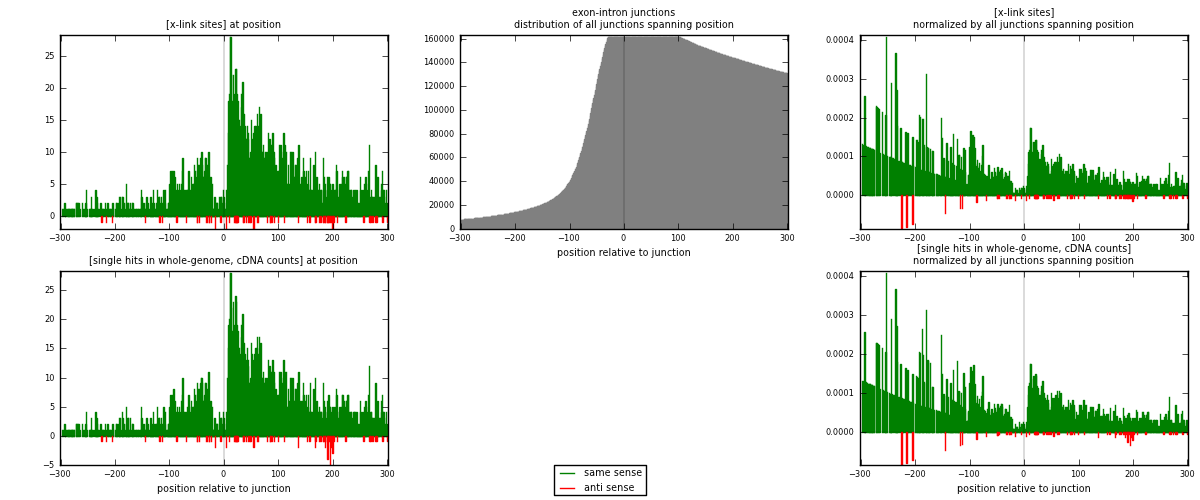 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2707 | 8.78% | 27.07% | 97.27% | 2476 | 1.07208 |
| anti | 76 | 0.25% | 0.76% | 2.73% | 52 | 0.030099 |
| both | 2783 | 9.03% | 27.83% | 100.00% | 2525 | 1.10218 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2764 | 8.59% | 26.48% | 96.91% | 2476 | 1.09465 |
| anti | 88 | 0.27% | 0.84% | 3.09% | 52 | 0.0348515 |
| both | 2852 | 8.86% | 27.32% | 100.00% | 2525 | 1.1295 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
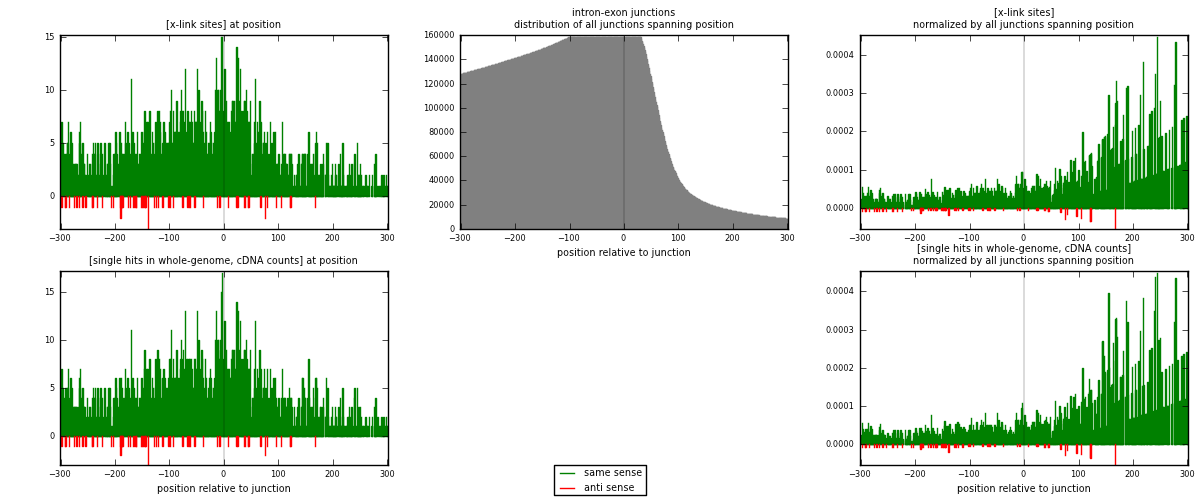 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2194 | 7.12% | 21.94% | 96.99% | 1979 | 1.07813 |
| anti | 68 | 0.22% | 0.68% | 3.01% | 57 | 0.0334152 |
| both | 2262 | 7.34% | 22.62% | 100.00% | 2035 | 1.11155 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2254 | 7.00% | 21.59% | 97.07% | 1979 | 1.10762 |
| anti | 68 | 0.21% | 0.65% | 2.93% | 57 | 0.0334152 |
| both | 2322 | 7.21% | 22.24% | 100.00% | 2035 | 1.14103 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
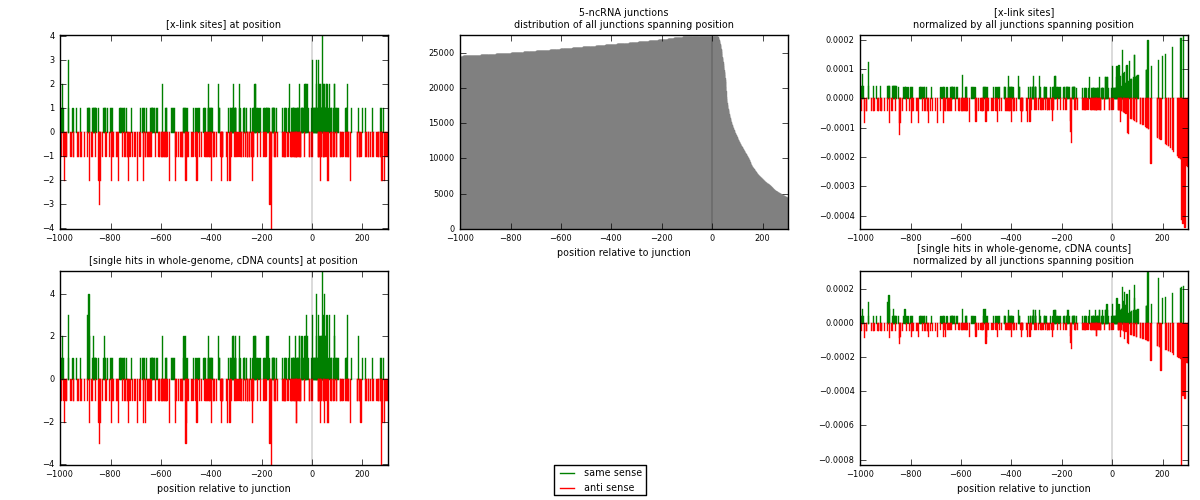 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 265 | 0.86% | 2.65% | 50.57% | 154 | 0.777126 |
| anti | 259 | 0.84% | 2.59% | 49.43% | 187 | 0.759531 |
| both | 524 | 1.70% | 5.24% | 100.00% | 341 | 1.53666 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 299 | 0.93% | 2.86% | 52.73% | 154 | 0.876833 |
| anti | 268 | 0.83% | 2.57% | 47.27% | 187 | 0.785924 |
| both | 567 | 1.76% | 5.43% | 100.00% | 341 | 1.66276 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
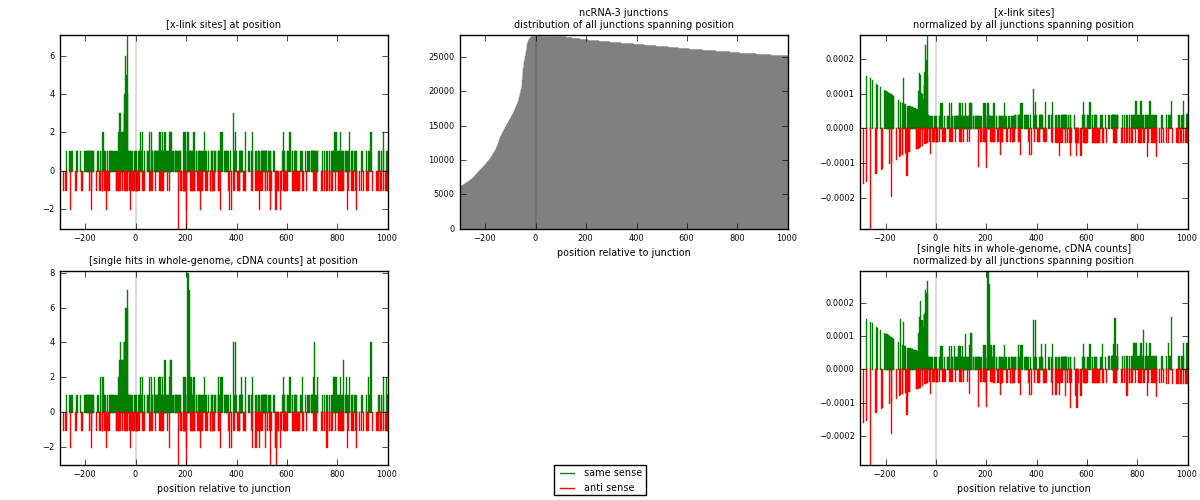 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 393 | 1.27% | 3.93% | 64.32% | 226 | 1.0155 |
| anti | 218 | 0.71% | 2.18% | 35.68% | 162 | 0.563307 |
| both | 611 | 1.98% | 6.11% | 100.00% | 387 | 1.57881 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 450 | 1.40% | 4.31% | 66.37% | 226 | 1.16279 |
| anti | 228 | 0.71% | 2.18% | 33.63% | 162 | 0.589147 |
| both | 678 | 2.11% | 6.49% | 100.00% | 387 | 1.75194 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
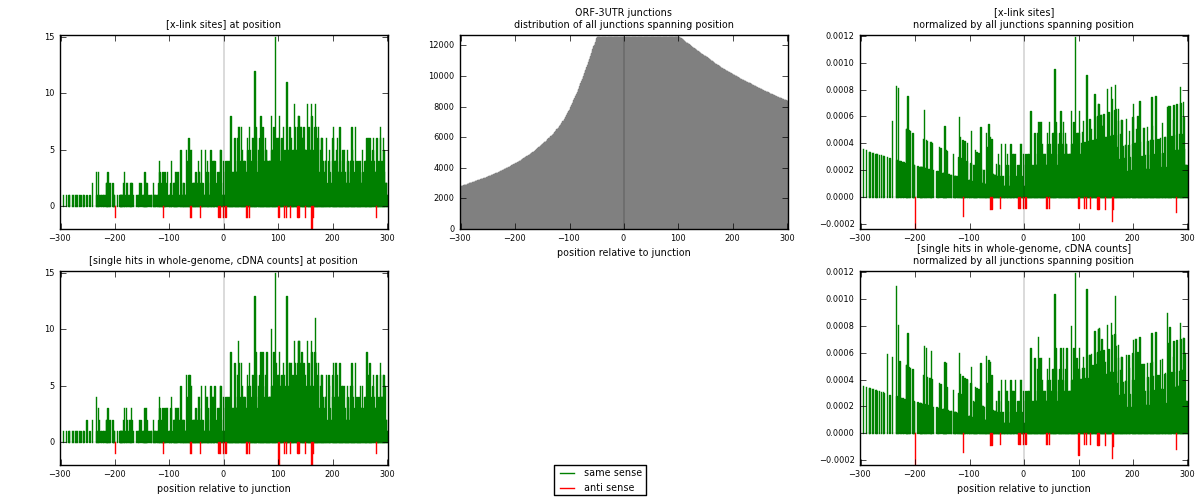 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1547 | 5.02% | 15.47% | 98.54% | 874 | 1.7343 |
| anti | 23 | 0.07% | 0.23% | 1.46% | 20 | 0.0257848 |
| both | 1570 | 5.09% | 15.70% | 100.00% | 892 | 1.76009 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1633 | 5.07% | 15.64% | 98.55% | 874 | 1.83072 |
| anti | 24 | 0.07% | 0.23% | 1.45% | 20 | 0.0269058 |
| both | 1657 | 5.15% | 15.87% | 100.00% | 892 | 1.85762 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
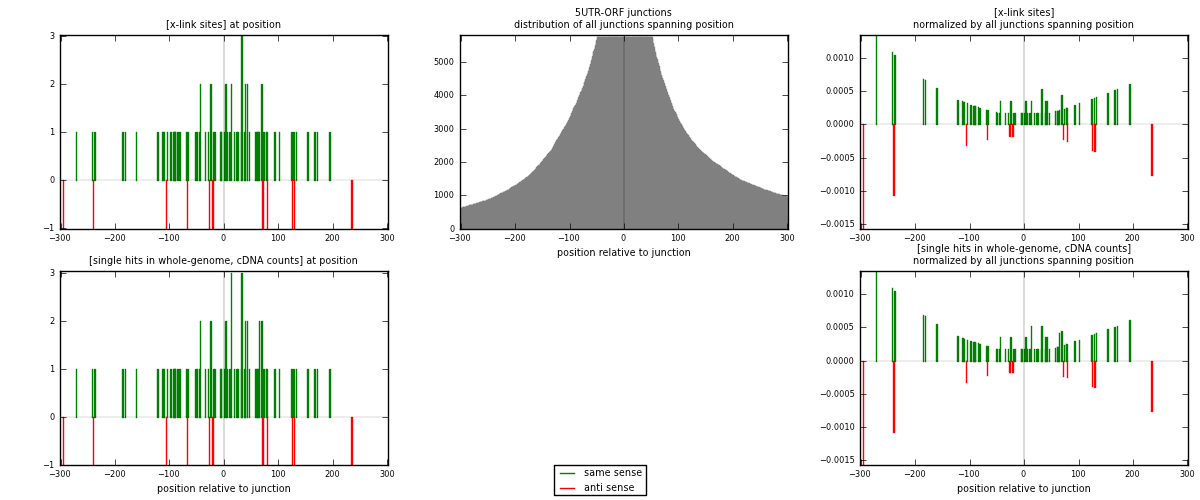 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 72 | 0.23% | 0.72% | 86.75% | 69 | 0.935065 |
| anti | 11 | 0.04% | 0.11% | 13.25% | 8 | 0.142857 |
| both | 83 | 0.27% | 0.83% | 100.00% | 77 | 1.07792 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 74 | 0.23% | 0.71% | 87.06% | 69 | 0.961039 |
| anti | 11 | 0.03% | 0.11% | 12.94% | 8 | 0.142857 |
| both | 85 | 0.26% | 0.81% | 100.00% | 77 | 1.1039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
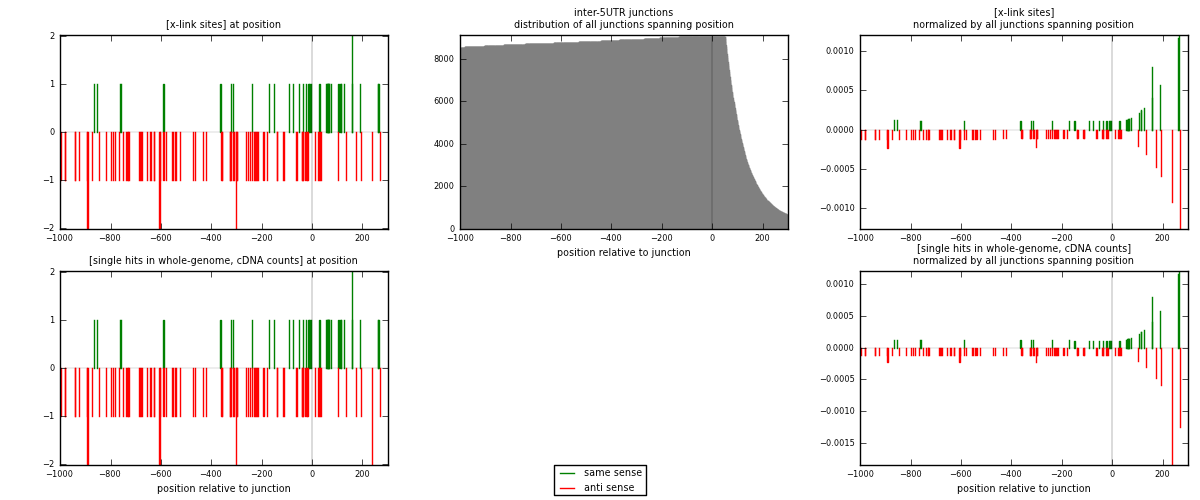 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 38 | 0.12% | 0.38% | 29.92% | 37 | 0.348624 |
| anti | 89 | 0.29% | 0.89% | 70.08% | 72 | 0.816514 |
| both | 127 | 0.41% | 1.27% | 100.00% | 109 | 1.16514 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 38 | 0.12% | 0.36% | 29.69% | 37 | 0.348624 |
| anti | 90 | 0.28% | 0.86% | 70.31% | 72 | 0.825688 |
| both | 128 | 0.40% | 1.23% | 100.00% | 109 | 1.17431 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
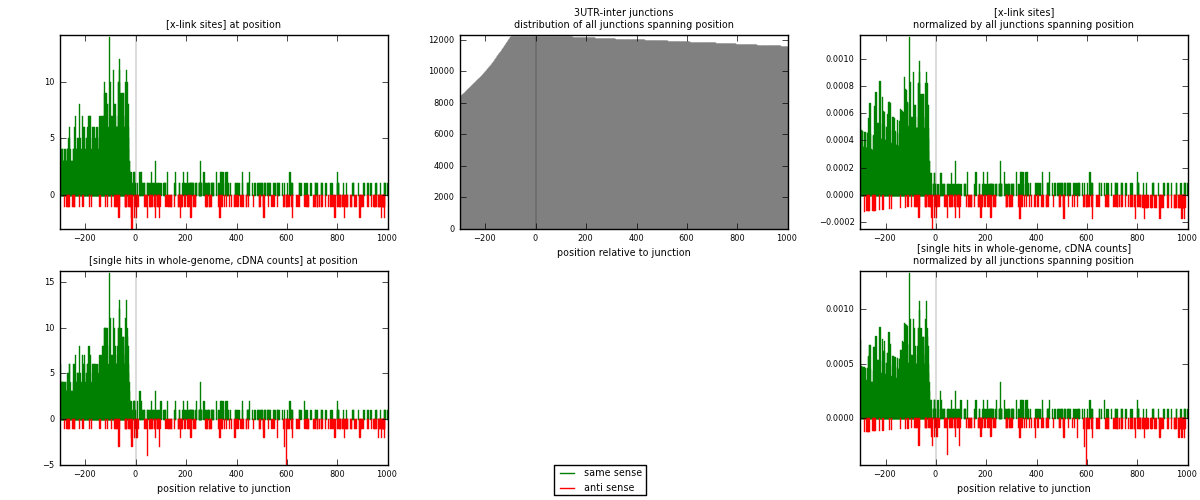 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1431 | 4.64% | 14.31% | 86.00% | 898 | 1.37596 |
| anti | 233 | 0.76% | 2.33% | 14.00% | 148 | 0.224038 |
| both | 1664 | 5.40% | 16.64% | 100.00% | 1040 | 1.6 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1501 | 4.66% | 14.38% | 85.87% | 898 | 1.44327 |
| anti | 247 | 0.77% | 2.37% | 14.13% | 148 | 0.2375 |
| both | 1748 | 5.43% | 16.74% | 100.00% | 1040 | 1.68077 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
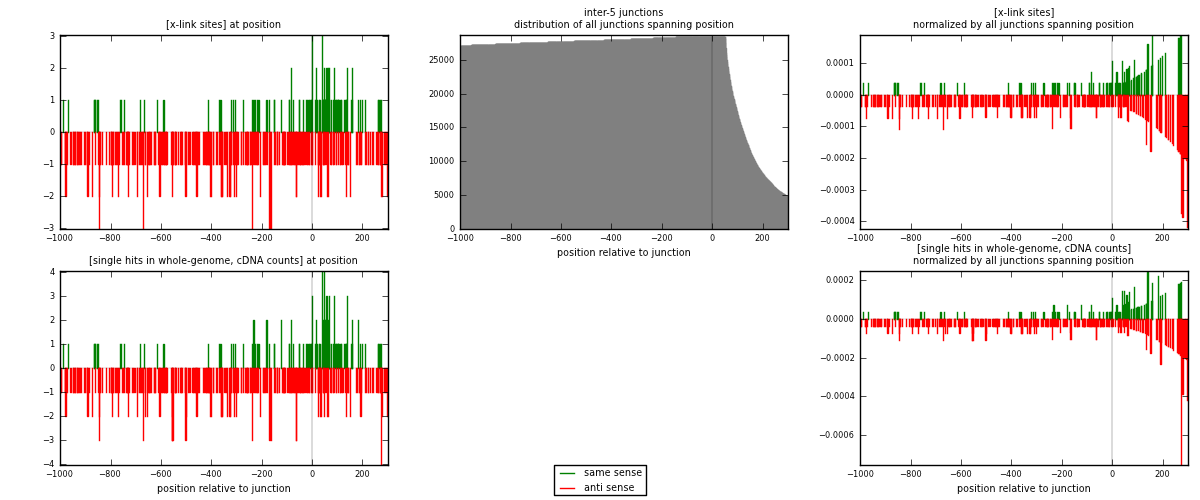
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 114 | 0.37% | 1.14% | 25.00% | 100 | 0.336283 |
| anti | 342 | 1.11% | 3.42% | 75.00% | 240 | 1.00885 |
| both | 456 | 1.48% | 4.56% | 100.00% | 339 | 1.34513 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 128 | 0.40% | 1.23% | 26.39% | 100 | 0.377581 |
| anti | 357 | 1.11% | 3.42% | 73.61% | 240 | 1.0531 |
| both | 485 | 1.51% | 4.65% | 100.00% | 339 | 1.43068 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
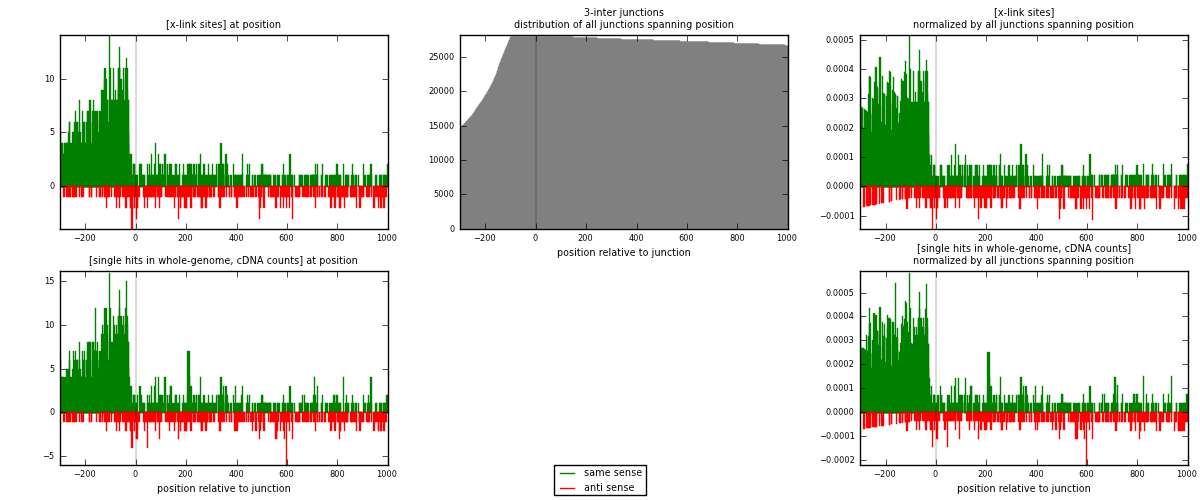
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1810 | 5.87% | 18.10% | 80.48% | 1090 | 1.32795 |
| anti | 439 | 1.42% | 4.39% | 19.52% | 282 | 0.322084 |
| both | 2249 | 7.30% | 22.49% | 100.00% | 1363 | 1.65004 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1963 | 6.10% | 18.80% | 81.02% | 1090 | 1.44021 |
| anti | 460 | 1.43% | 4.41% | 18.98% | 282 | 0.337491 |
| both | 2423 | 7.53% | 23.21% | 100.00% | 1363 | 1.7777 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
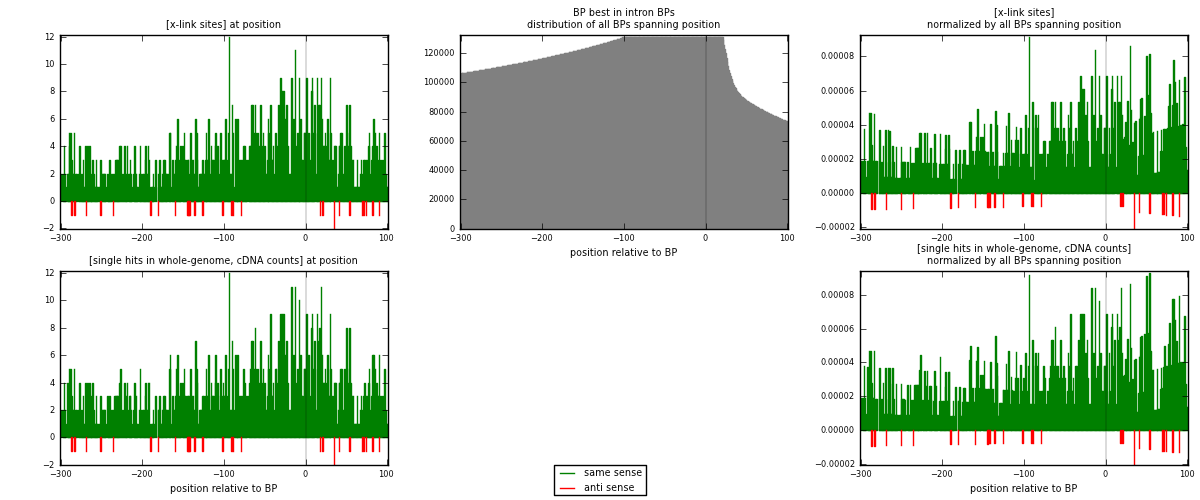
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 1186 | 3.85% | 11.86% | 97.61% | 1117 | 1.03762 |
| anti | 29 | 0.09% | 0.29% | 2.39% | 26 | 0.0253718 |
| both | 1215 | 3.94% | 12.15% | 100.00% | 1143 | 1.06299 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 1232 | 3.83% | 11.80% | 97.70% | 1117 | 1.07787 |
| anti | 29 | 0.09% | 0.28% | 2.30% | 26 | 0.0253718 |
| both | 1261 | 3.92% | 12.08% | 100.00% | 1143 | 1.10324 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.