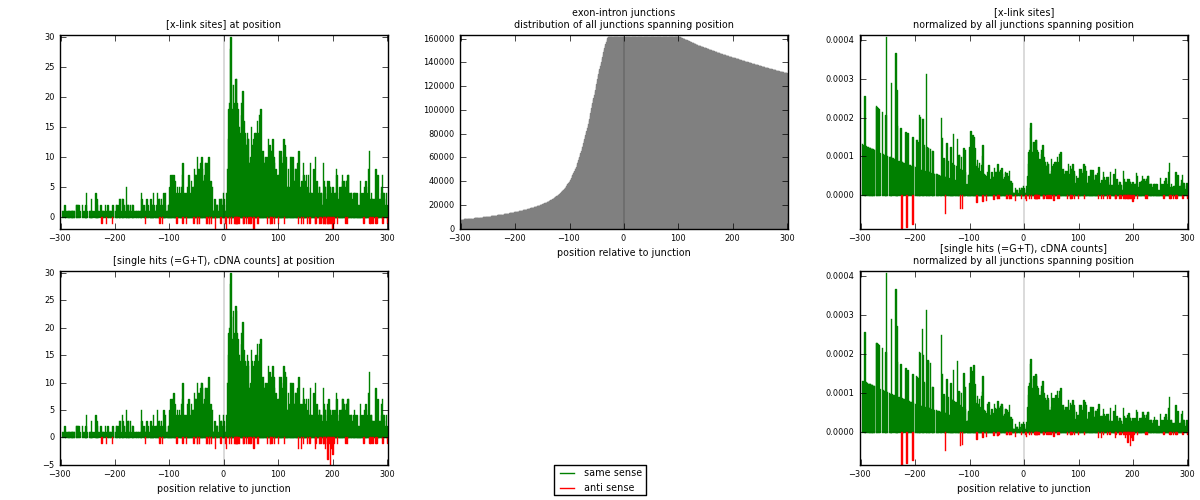
RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2739 | 8.52% | 26.50% | 96.99% | 2502 | 1.07076 |
| anti | 85 | 0.26% | 0.82% | 3.01% | 59 | 0.0332291 |
| both | 2824 | 8.78% | 27.32% | 100.00% | 2558 | 1.10399 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2798 | 8.30% | 25.85% | 96.62% | 2502 | 1.09382 |
| anti | 98 | 0.29% | 0.91% | 3.38% | 59 | 0.0383112 |
| both | 2896 | 8.59% | 26.76% | 100.00% | 2558 | 1.13213 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
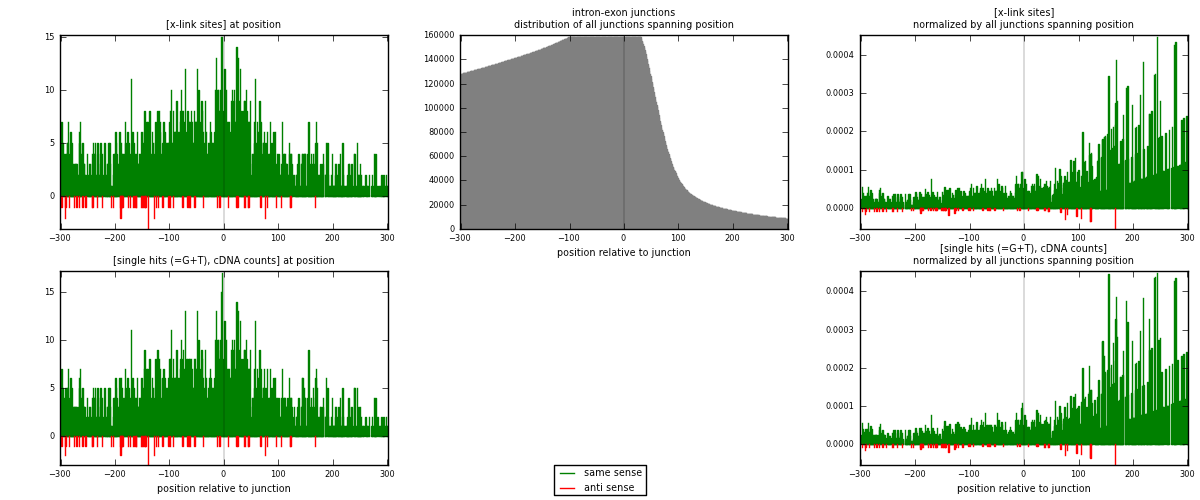
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2225 | 6.92% | 21.53% | 96.95% | 1997 | 1.08273 |
| anti | 70 | 0.22% | 0.68% | 3.05% | 59 | 0.0340633 |
| both | 2295 | 7.14% | 22.20% | 100.00% | 2055 | 1.11679 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2287 | 6.79% | 21.13% | 97.03% | 1997 | 1.1129 |
| anti | 70 | 0.21% | 0.65% | 2.97% | 59 | 0.0340633 |
| both | 2357 | 6.99% | 21.78% | 100.00% | 2055 | 1.14696 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
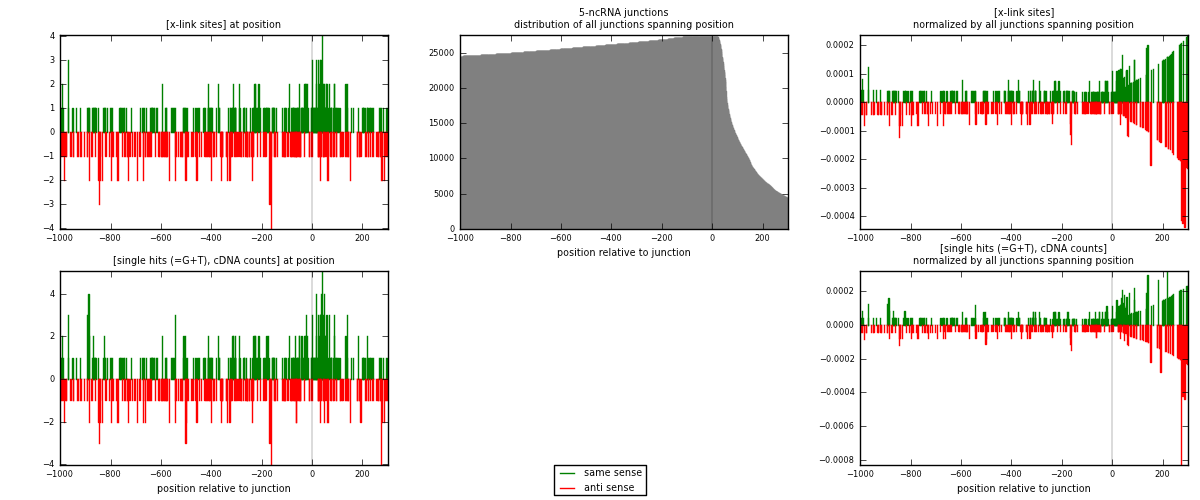
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 298 | 0.93% | 2.88% | 52.65% | 166 | 0.823204 |
| anti | 268 | 0.83% | 2.59% | 47.35% | 196 | 0.740331 |
| both | 566 | 1.76% | 5.48% | 100.00% | 362 | 1.56354 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 337 | 1.00% | 3.11% | 54.89% | 166 | 0.930939 |
| anti | 277 | 0.82% | 2.56% | 45.11% | 196 | 0.765193 |
| both | 614 | 1.82% | 5.67% | 100.00% | 362 | 1.69613 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
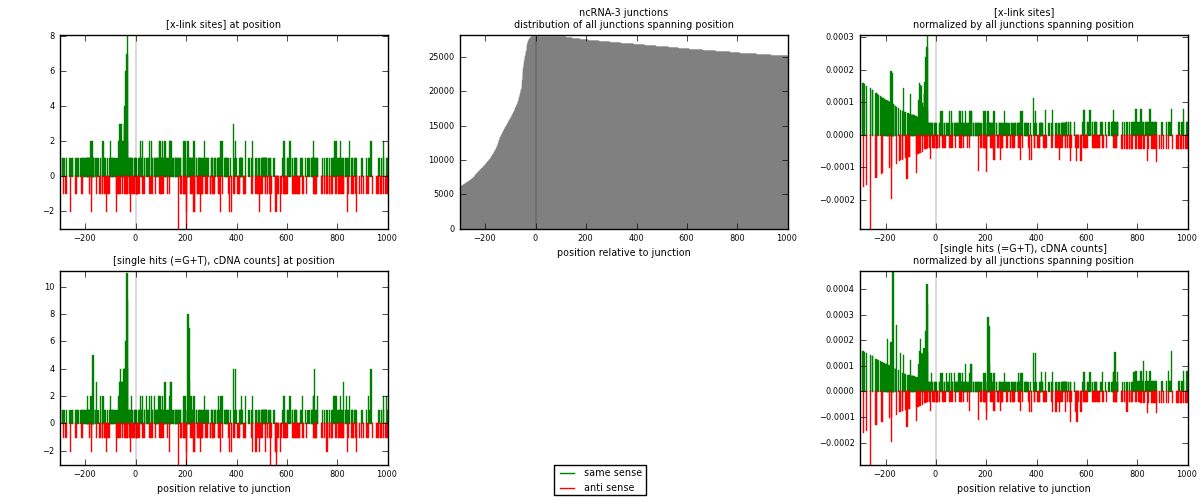
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 438 | 1.36% | 4.24% | 65.77% | 237 | 1.08685 |
| anti | 228 | 0.71% | 2.21% | 34.23% | 168 | 0.565757 |
| both | 666 | 2.07% | 6.44% | 100.00% | 403 | 1.65261 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 509 | 1.51% | 4.70% | 68.14% | 237 | 1.26303 |
| anti | 238 | 0.71% | 2.20% | 31.86% | 168 | 0.590571 |
| both | 747 | 2.22% | 6.90% | 100.00% | 403 | 1.8536 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
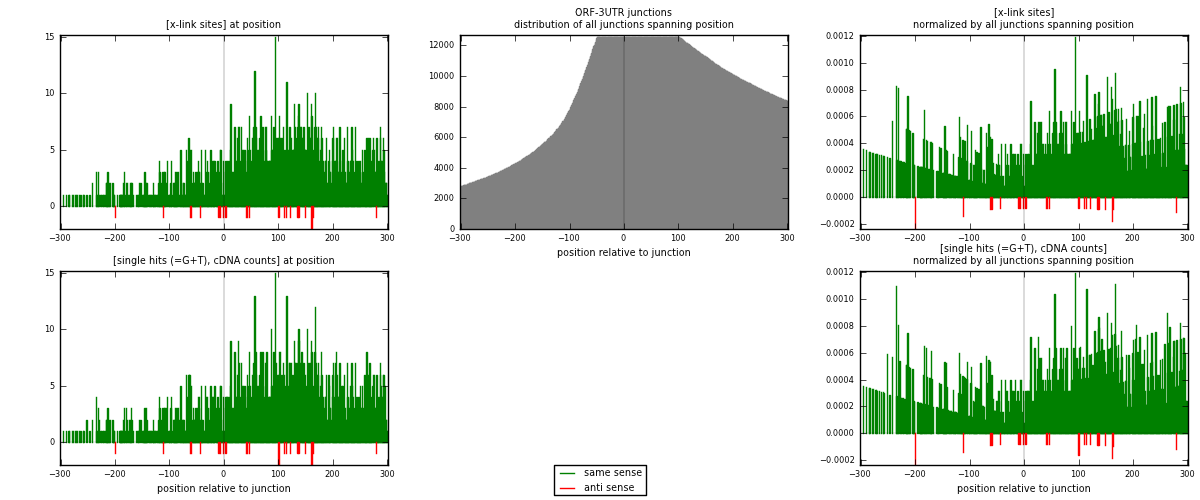
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1613 | 5.02% | 15.61% | 98.59% | 898 | 1.76092 |
| anti | 23 | 0.07% | 0.22% | 1.41% | 20 | 0.0251092 |
| both | 1636 | 5.09% | 15.83% | 100.00% | 916 | 1.78603 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1704 | 5.06% | 15.75% | 98.61% | 898 | 1.86026 |
| anti | 24 | 0.07% | 0.22% | 1.39% | 20 | 0.0262009 |
| both | 1728 | 5.13% | 15.97% | 100.00% | 916 | 1.88646 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
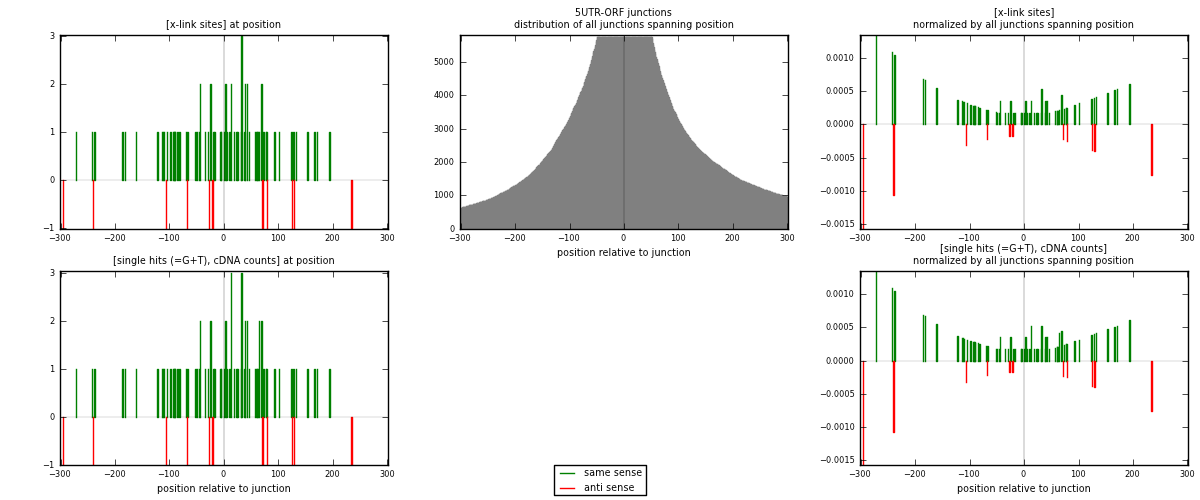
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 72 | 0.22% | 0.70% | 86.75% | 69 | 0.935065 |
| anti | 11 | 0.03% | 0.11% | 13.25% | 8 | 0.142857 |
| both | 83 | 0.26% | 0.80% | 100.00% | 77 | 1.07792 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 74 | 0.22% | 0.68% | 87.06% | 69 | 0.961039 |
| anti | 11 | 0.03% | 0.10% | 12.94% | 8 | 0.142857 |
| both | 85 | 0.25% | 0.79% | 100.00% | 77 | 1.1039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
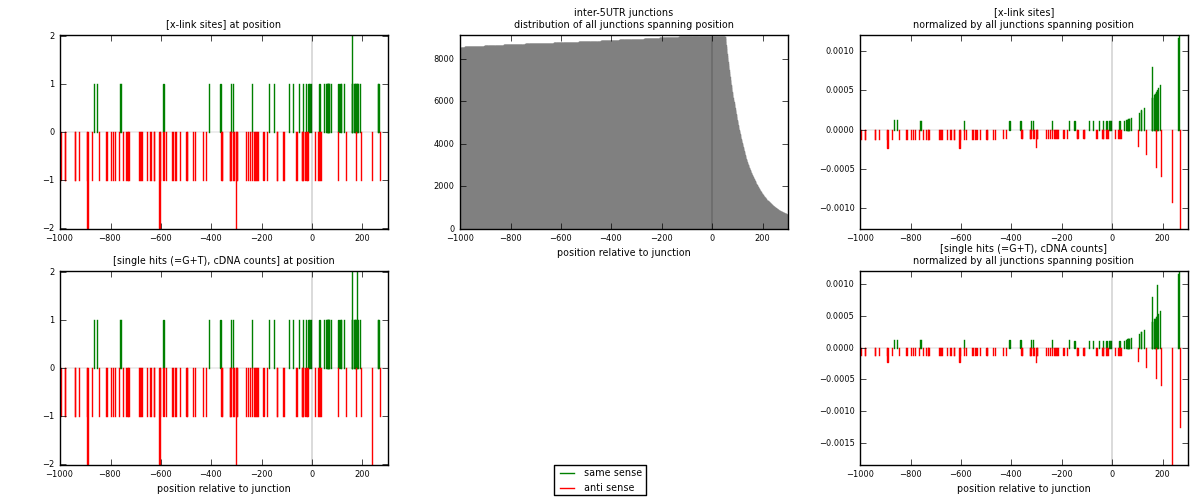
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 48 | 0.15% | 0.46% | 34.29% | 39 | 0.428571 |
| anti | 92 | 0.29% | 0.89% | 65.71% | 73 | 0.821429 |
| both | 140 | 0.44% | 1.35% | 100.00% | 112 | 1.25 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 49 | 0.15% | 0.45% | 34.51% | 39 | 0.4375 |
| anti | 93 | 0.28% | 0.86% | 65.49% | 73 | 0.830357 |
| both | 142 | 0.42% | 1.31% | 100.00% | 112 | 1.26786 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
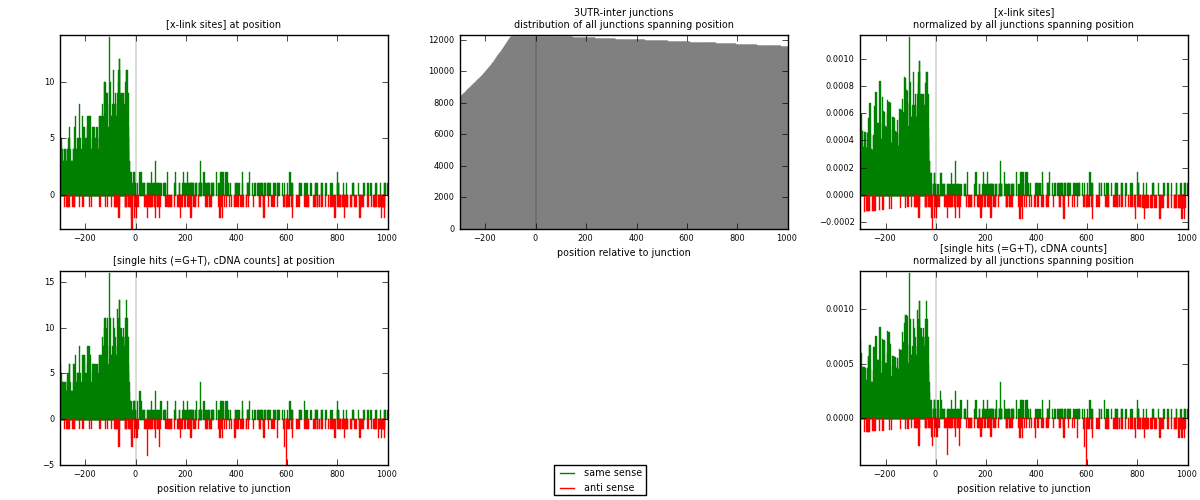
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1502 | 4.67% | 14.53% | 86.27% | 921 | 1.40901 |
| anti | 239 | 0.74% | 2.31% | 13.73% | 151 | 0.224203 |
| both | 1741 | 5.41% | 16.84% | 100.00% | 1066 | 1.63321 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1575 | 4.67% | 14.55% | 86.16% | 921 | 1.47749 |
| anti | 253 | 0.75% | 2.34% | 13.84% | 151 | 0.237336 |
| both | 1828 | 5.42% | 16.89% | 100.00% | 1066 | 1.71482 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
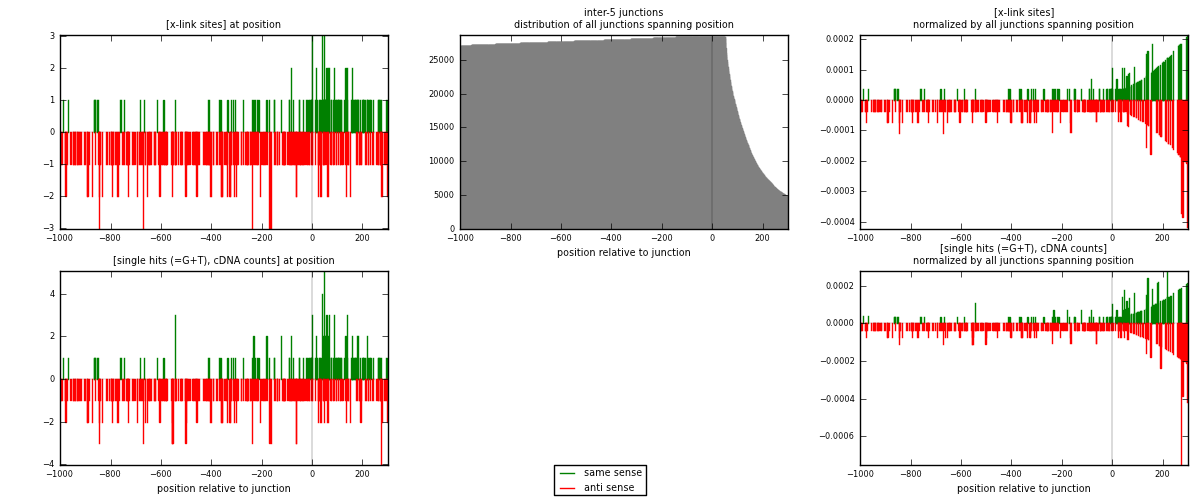
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 147 | 0.46% | 1.42% | 29.46% | 108 | 0.415254 |
| anti | 352 | 1.09% | 3.41% | 70.54% | 247 | 0.99435 |
| both | 499 | 1.55% | 4.83% | 100.00% | 354 | 1.4096 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 165 | 0.49% | 1.52% | 31.02% | 108 | 0.466102 |
| anti | 367 | 1.09% | 3.39% | 68.98% | 247 | 1.03672 |
| both | 532 | 1.58% | 4.92% | 100.00% | 354 | 1.50282 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
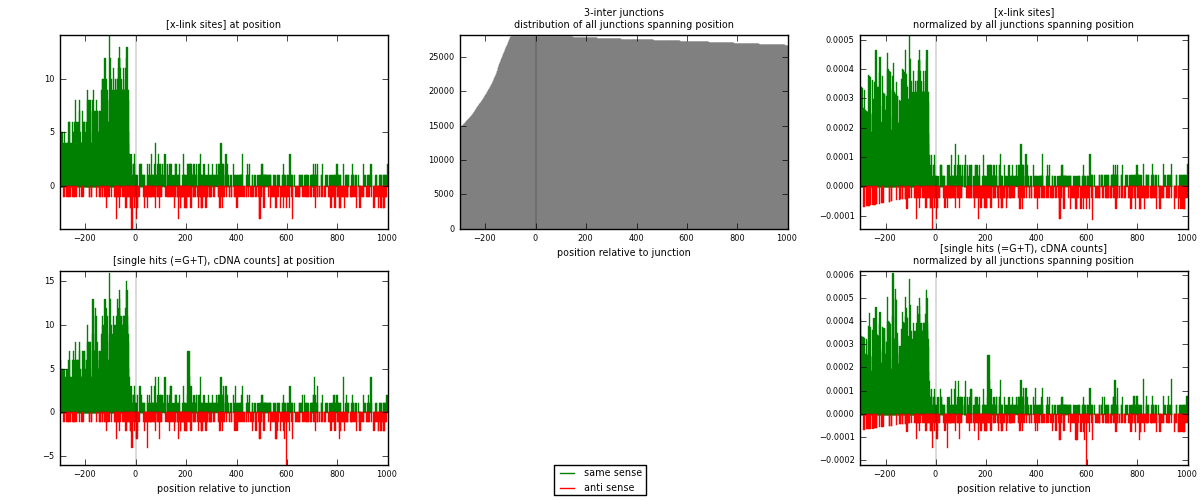
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1924 | 5.98% | 18.61% | 81.01% | 1122 | 1.37429 |
| anti | 451 | 1.40% | 4.36% | 18.99% | 289 | 0.322143 |
| both | 2375 | 7.39% | 22.98% | 100.00% | 1400 | 1.69643 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2100 | 6.23% | 19.40% | 81.65% | 1122 | 1.5 |
| anti | 472 | 1.40% | 4.36% | 18.35% | 289 | 0.337143 |
| both | 2572 | 7.63% | 23.77% | 100.00% | 1400 | 1.83714 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
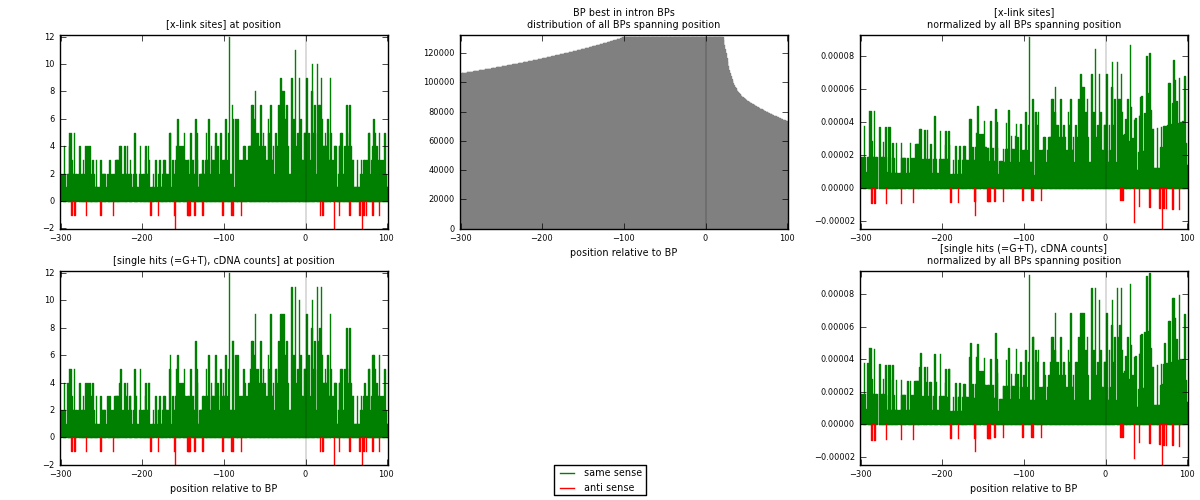
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 1202 | 3.74% | 11.63% | 97.33% | 1131 | 1.03621 |
| anti | 33 | 0.10% | 0.32% | 2.67% | 29 | 0.0284483 |
| both | 1235 | 3.84% | 11.95% | 100.00% | 1160 | 1.06466 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 1250 | 3.71% | 11.55% | 97.43% | 1131 | 1.07759 |
| anti | 33 | 0.10% | 0.30% | 2.57% | 29 | 0.0284483 |
| both | 1283 | 3.81% | 11.86% | 100.00% | 1160 | 1.10603 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.