RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
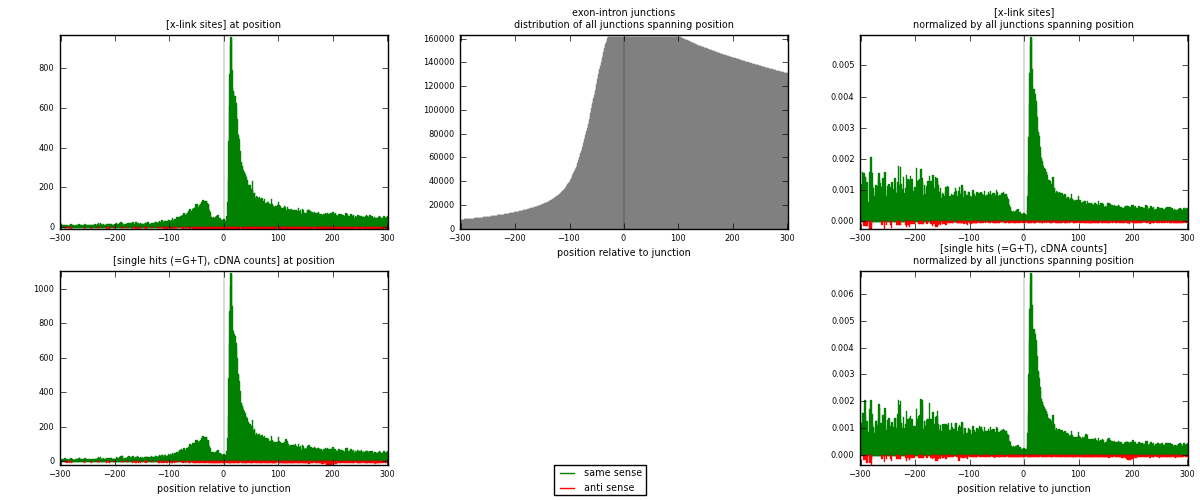 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 48584 | 10.93% | 34.84% | 98.00% | 28798 | 1.65252 |
| anti | 992 | 0.22% | 0.71% | 2.00% | 718 | 0.0337415 |
| both | 49576 | 11.15% | 35.55% | 100.00% | 29400 | 1.68626 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 51786 | 10.54% | 32.42% | 97.63% | 28798 | 1.76143 |
| anti | 1255 | 0.26% | 0.79% | 2.37% | 718 | 0.0426871 |
| both | 53041 | 10.80% | 33.20% | 100.00% | 29400 | 1.80412 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
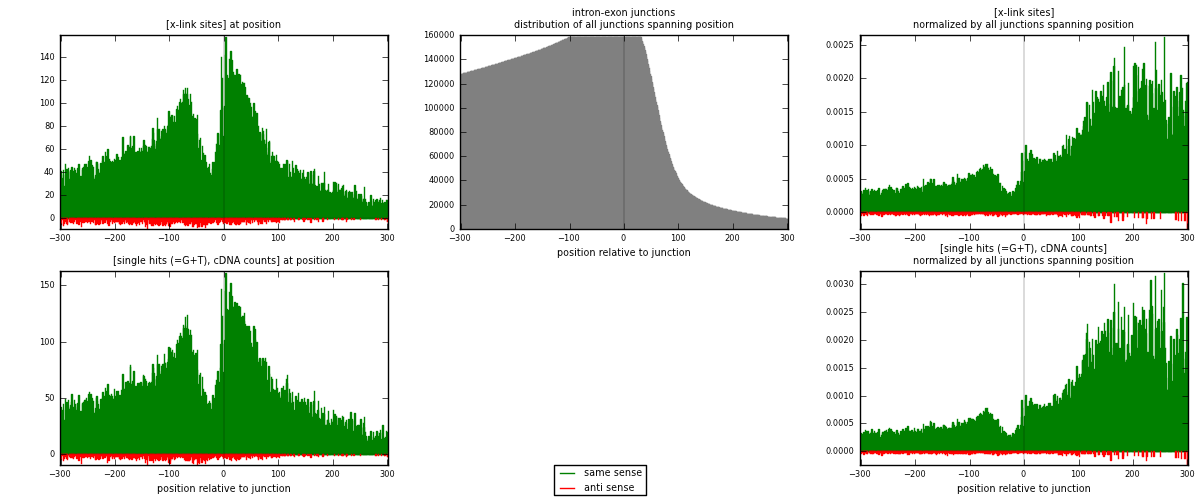 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 31761 | 7.15% | 22.78% | 96.79% | 20667 | 1.48631 |
| anti | 1053 | 0.24% | 0.76% | 3.21% | 796 | 0.049277 |
| both | 32814 | 7.38% | 23.53% | 100.00% | 21369 | 1.53559 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 34048 | 6.93% | 21.31% | 96.85% | 20667 | 1.59334 |
| anti | 1108 | 0.23% | 0.69% | 3.15% | 796 | 0.0518508 |
| both | 35156 | 7.16% | 22.01% | 100.00% | 21369 | 1.64519 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
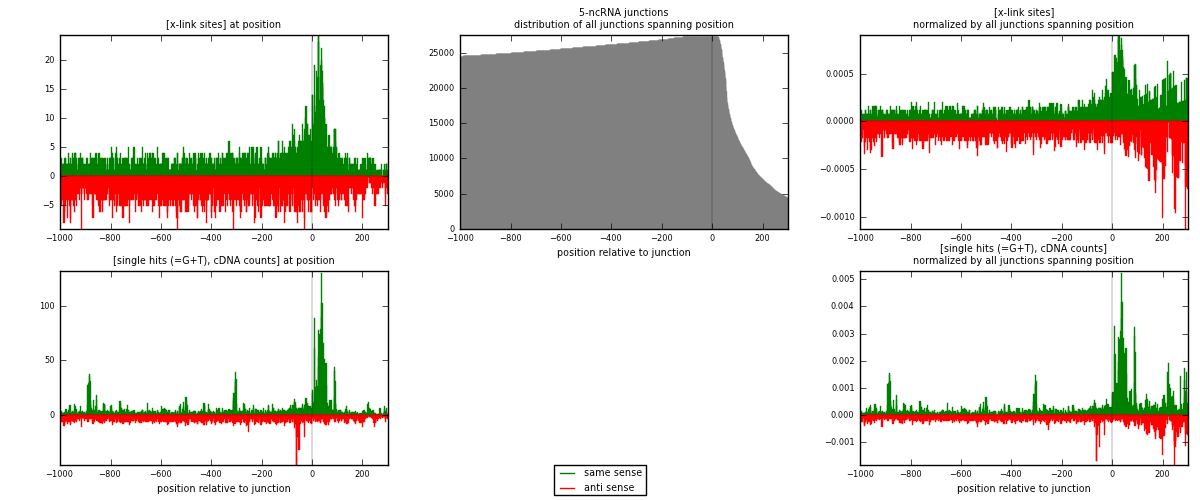 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3142 | 0.71% | 2.25% | 46.95% | 1292 | 1.09975 |
| anti | 3550 | 0.80% | 2.55% | 53.05% | 1623 | 1.24256 |
| both | 6692 | 1.51% | 4.80% | 100.00% | 2857 | 2.34232 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 6063 | 1.23% | 3.80% | 60.14% | 1292 | 2.12216 |
| anti | 4019 | 0.82% | 2.52% | 39.86% | 1623 | 1.40672 |
| both | 10082 | 2.05% | 6.31% | 100.00% | 2857 | 3.52888 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
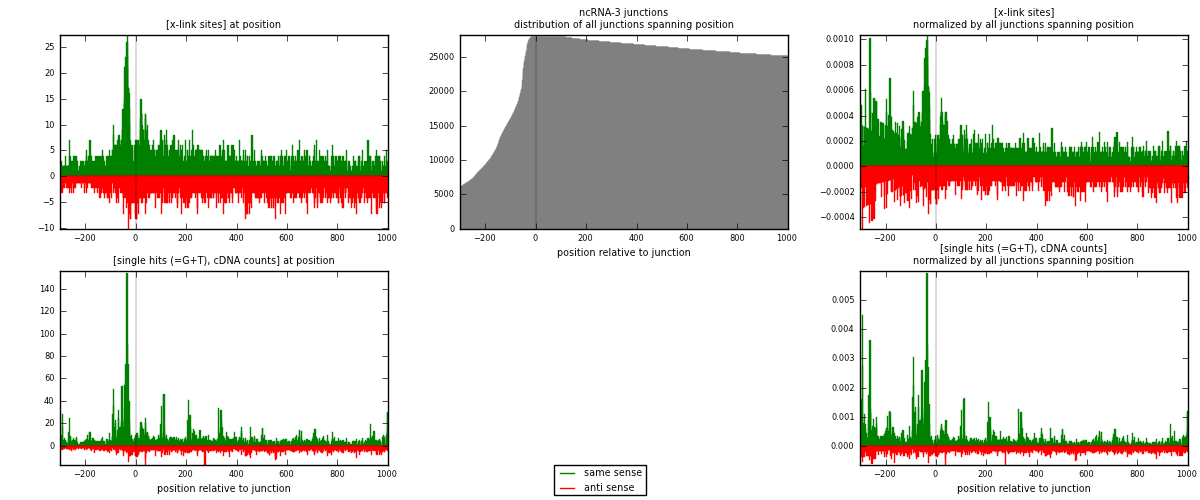 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3860 | 0.87% | 2.77% | 55.56% | 1569 | 1.31203 |
| anti | 3088 | 0.69% | 2.21% | 44.44% | 1420 | 1.04963 |
| both | 6948 | 1.56% | 4.98% | 100.00% | 2942 | 2.36166 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 6869 | 1.40% | 4.30% | 67.00% | 1569 | 2.33481 |
| anti | 3383 | 0.69% | 2.12% | 33.00% | 1420 | 1.1499 |
| both | 10252 | 2.09% | 6.42% | 100.00% | 2942 | 3.4847 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
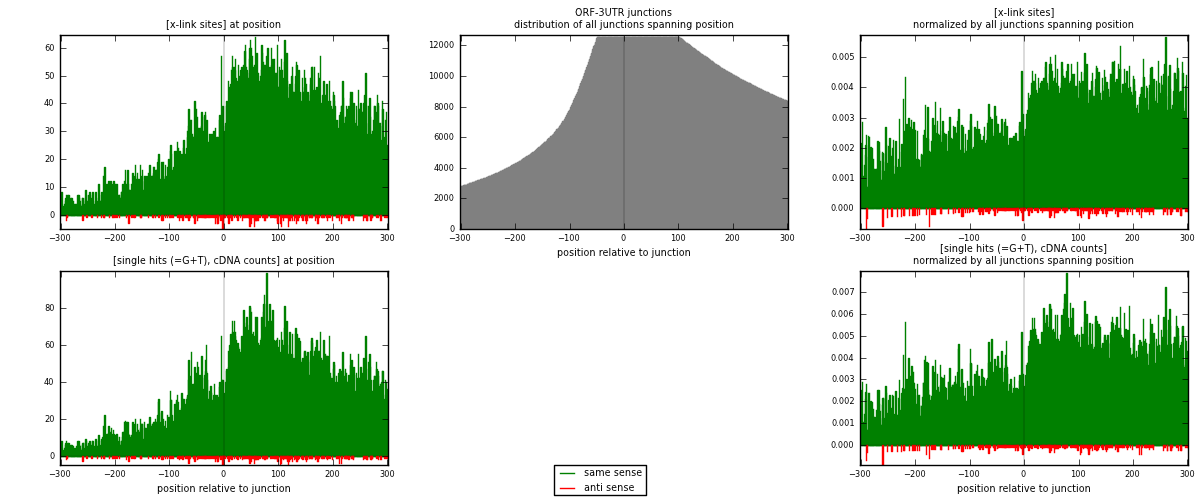
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 17254 | 3.88% | 12.37% | 98.27% | 4148 | 4.02943 |
| anti | 303 | 0.07% | 0.22% | 1.73% | 198 | 0.0707613 |
| both | 17557 | 3.95% | 12.59% | 100.00% | 4282 | 4.10019 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 21075 | 4.29% | 13.19% | 98.50% | 4148 | 4.92177 |
| anti | 322 | 0.07% | 0.20% | 1.50% | 198 | 0.0751985 |
| both | 21397 | 4.36% | 13.40% | 100.00% | 4282 | 4.99696 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
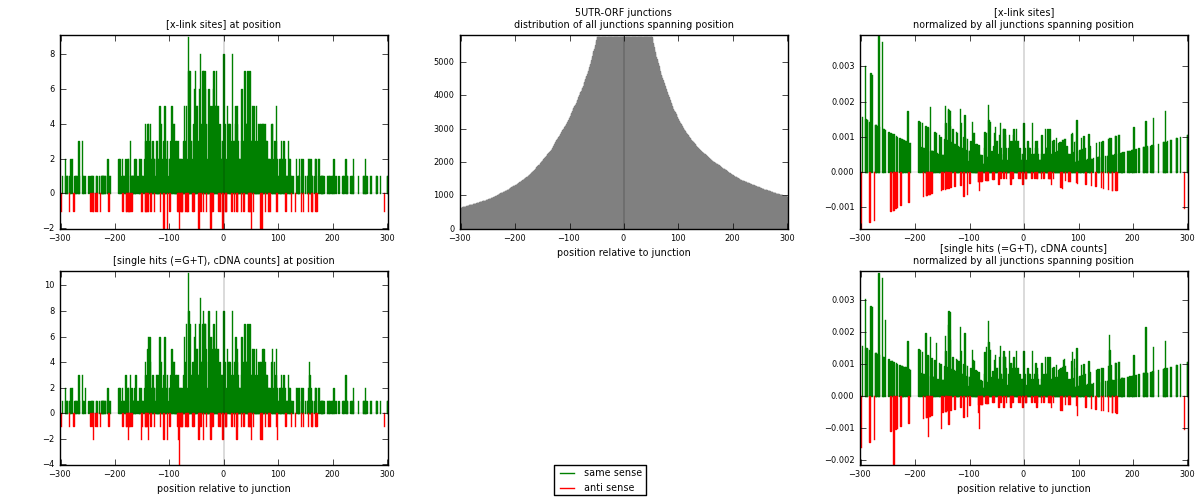
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 888 | 0.20% | 0.64% | 89.88% | 551 | 1.50254 |
| anti | 100 | 0.02% | 0.07% | 10.12% | 44 | 0.169205 |
| both | 988 | 0.22% | 0.71% | 100.00% | 591 | 1.67174 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 941 | 0.19% | 0.59% | 89.53% | 551 | 1.59222 |
| anti | 110 | 0.02% | 0.07% | 10.47% | 44 | 0.186125 |
| both | 1051 | 0.21% | 0.66% | 100.00% | 591 | 1.77834 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
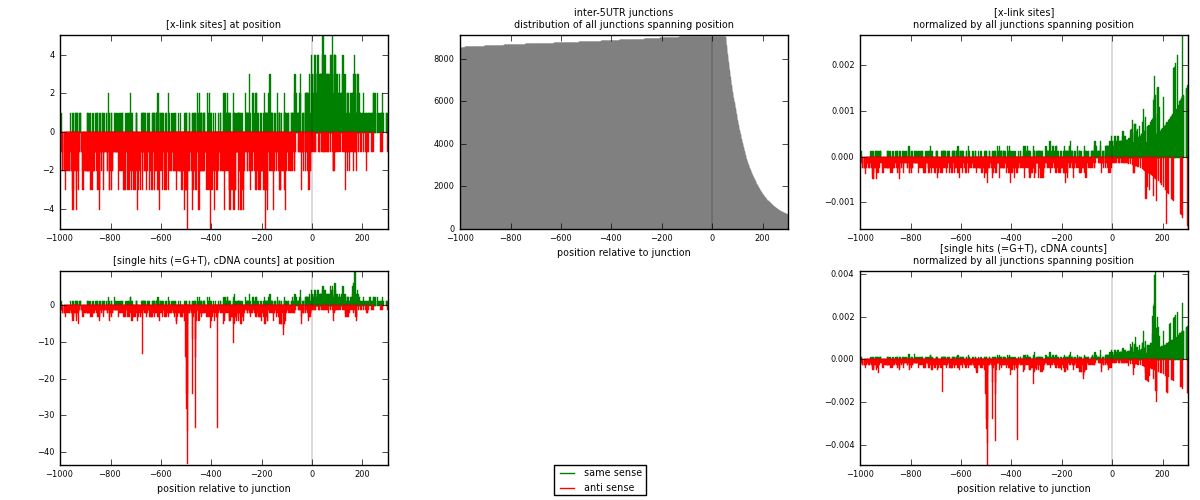
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 665 | 0.15% | 0.48% | 35.22% | 471 | 0.607306 |
| anti | 1223 | 0.28% | 0.88% | 64.78% | 682 | 1.11689 |
| both | 1888 | 0.42% | 1.35% | 100.00% | 1095 | 1.7242 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 718 | 0.15% | 0.45% | 30.71% | 471 | 0.655708 |
| anti | 1620 | 0.33% | 1.01% | 69.29% | 682 | 1.47945 |
| both | 2338 | 0.48% | 1.46% | 100.00% | 1095 | 2.13516 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
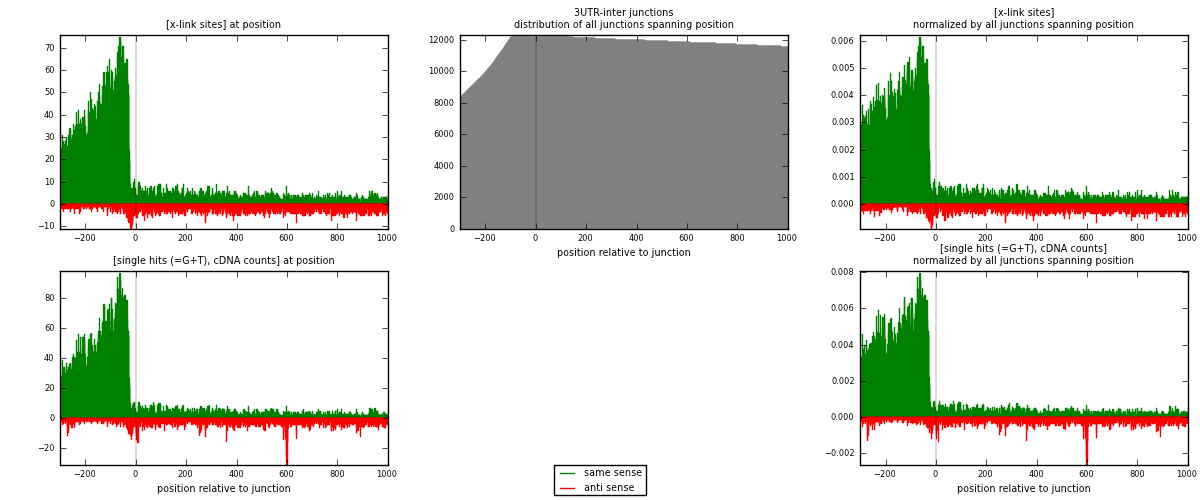
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13994 | 3.15% | 10.04% | 83.41% | 3951 | 3.09602 |
| anti | 2784 | 0.63% | 2.00% | 16.59% | 864 | 0.615929 |
| both | 16778 | 3.77% | 12.03% | 100.00% | 4520 | 3.71195 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 16504 | 3.36% | 10.33% | 82.69% | 3951 | 3.65133 |
| anti | 3456 | 0.70% | 2.16% | 17.31% | 864 | 0.764602 |
| both | 19960 | 4.06% | 12.50% | 100.00% | 4520 | 4.41593 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
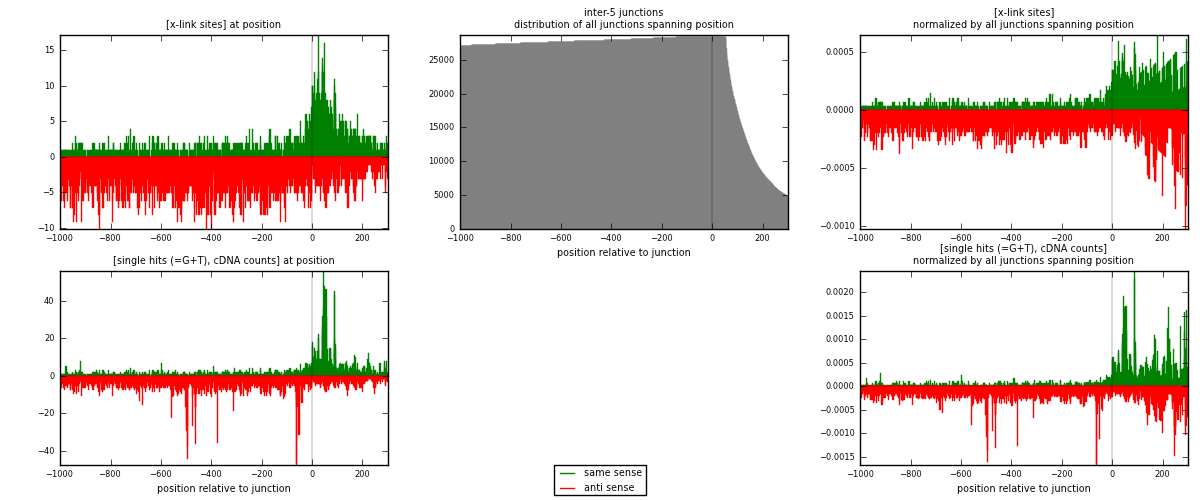
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1818 | 0.41% | 1.30% | 29.78% | 1019 | 0.610682 |
| anti | 4286 | 0.96% | 3.07% | 70.22% | 2068 | 1.4397 |
| both | 6104 | 1.37% | 4.38% | 100.00% | 2977 | 2.05039 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2589 | 0.53% | 1.62% | 33.31% | 1019 | 0.869667 |
| anti | 5184 | 1.06% | 3.25% | 66.69% | 2068 | 1.74135 |
| both | 7773 | 1.58% | 4.87% | 100.00% | 2977 | 2.61102 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
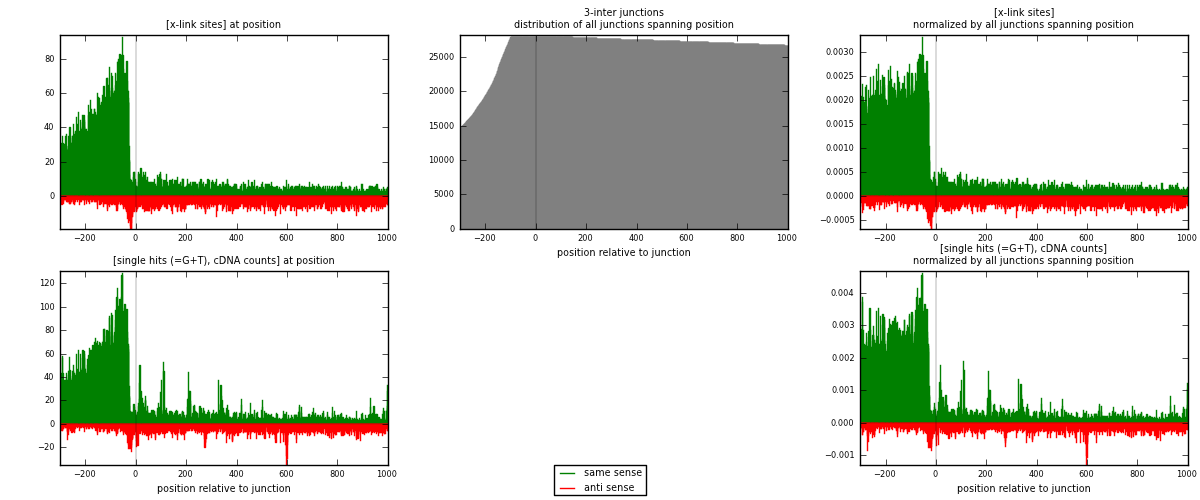
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 17150 | 3.86% | 12.30% | 76.05% | 4896 | 2.68556 |
| anti | 5402 | 1.22% | 3.87% | 23.95% | 1849 | 0.845913 |
| both | 22552 | 5.07% | 16.17% | 100.00% | 6386 | 3.53148 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 22349 | 4.55% | 13.99% | 77.59% | 4896 | 3.49969 |
| anti | 6456 | 1.31% | 4.04% | 22.41% | 1849 | 1.01096 |
| both | 28805 | 5.87% | 18.03% | 100.00% | 6386 | 4.51065 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
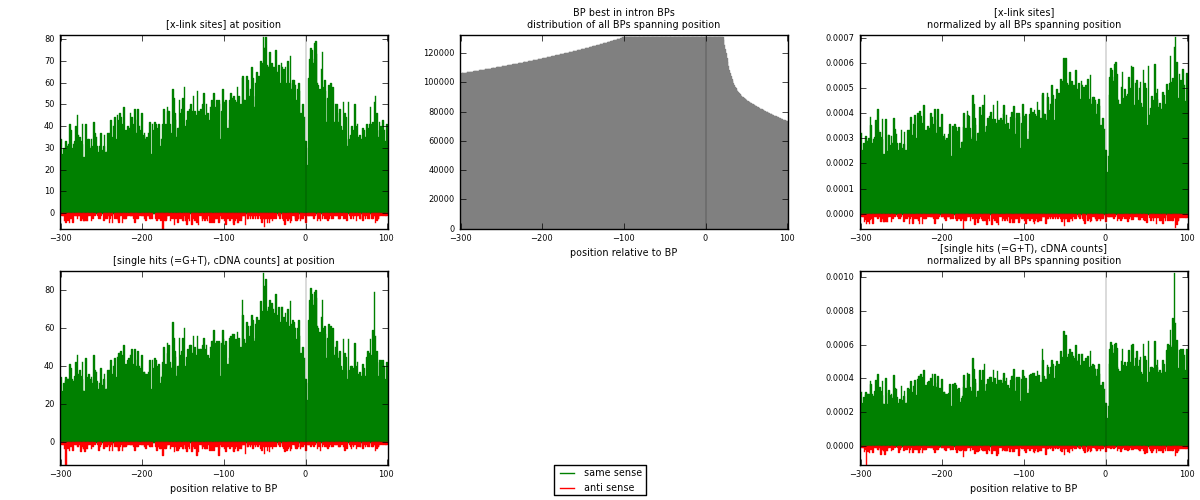
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 17684 | 3.98% | 12.68% | 96.49% | 13050 | 1.30664 |
| anti | 644 | 0.14% | 0.46% | 3.51% | 521 | 0.0475839 |
| both | 18328 | 4.12% | 13.14% | 100.00% | 13534 | 1.35422 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 18413 | 3.75% | 11.53% | 96.39% | 13050 | 1.3605 |
| anti | 690 | 0.14% | 0.43% | 3.61% | 521 | 0.0509827 |
| both | 19103 | 3.89% | 11.96% | 100.00% | 13534 | 1.41148 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.