RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
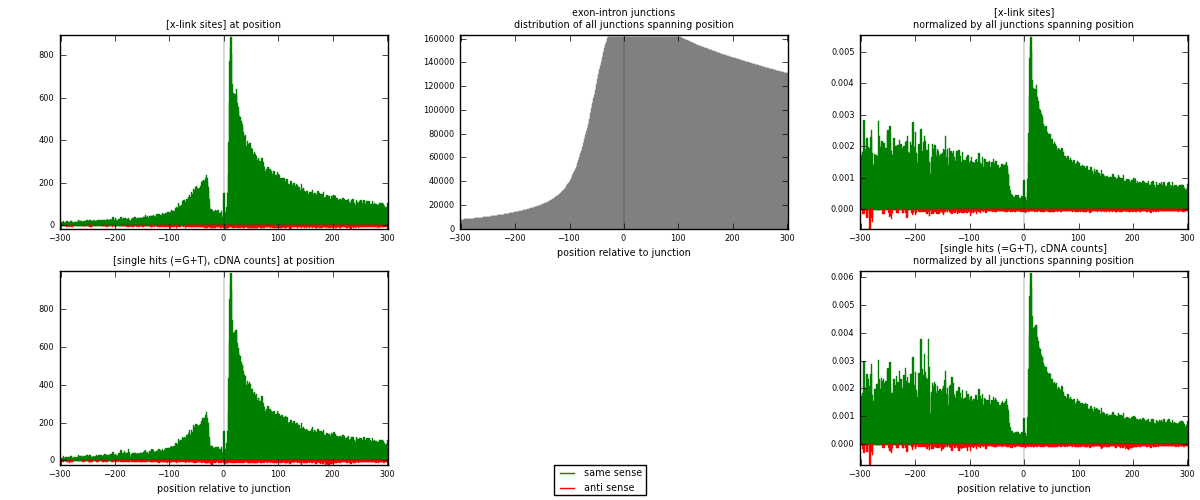 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 79855 | 9.84% | 32.77% | 97.83% | 39268 | 1.98689 |
| anti | 1768 | 0.22% | 0.73% | 2.17% | 1168 | 0.0439899 |
| both | 81623 | 10.06% | 33.50% | 100.00% | 40191 | 2.03088 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 84653 | 9.36% | 30.10% | 97.52% | 39268 | 2.10627 |
| anti | 2153 | 0.24% | 0.77% | 2.48% | 1168 | 0.0535692 |
| both | 86806 | 9.60% | 30.87% | 100.00% | 40191 | 2.15984 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
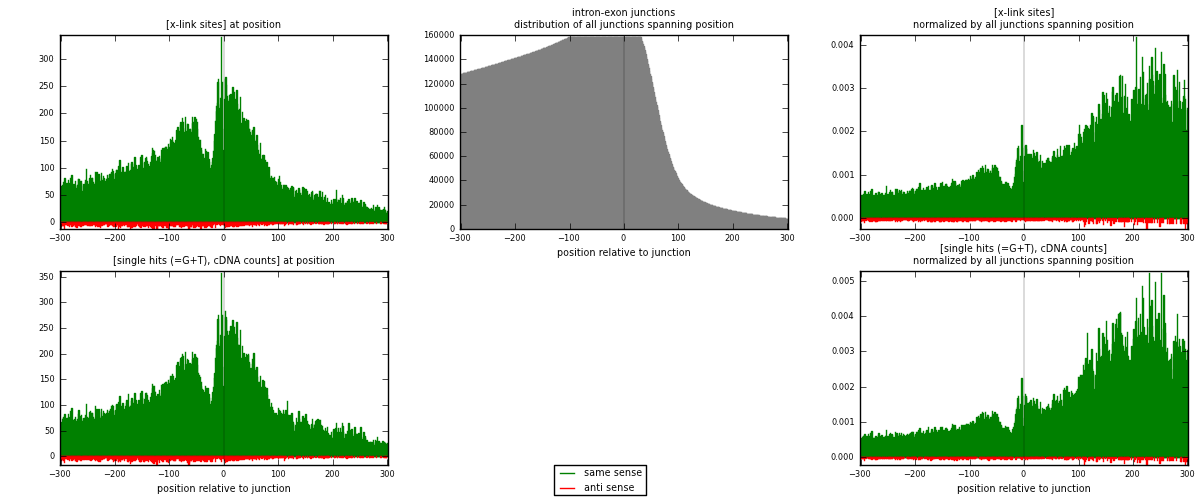 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 57572 | 7.09% | 23.63% | 96.66% | 31950 | 1.74081 |
| anti | 1991 | 0.25% | 0.82% | 3.34% | 1393 | 0.060202 |
| both | 59563 | 7.34% | 24.45% | 100.00% | 33072 | 1.80101 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 61791 | 6.83% | 21.97% | 96.65% | 31950 | 1.86838 |
| anti | 2144 | 0.24% | 0.76% | 3.35% | 1393 | 0.0648283 |
| both | 63935 | 7.07% | 22.73% | 100.00% | 33072 | 1.93321 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
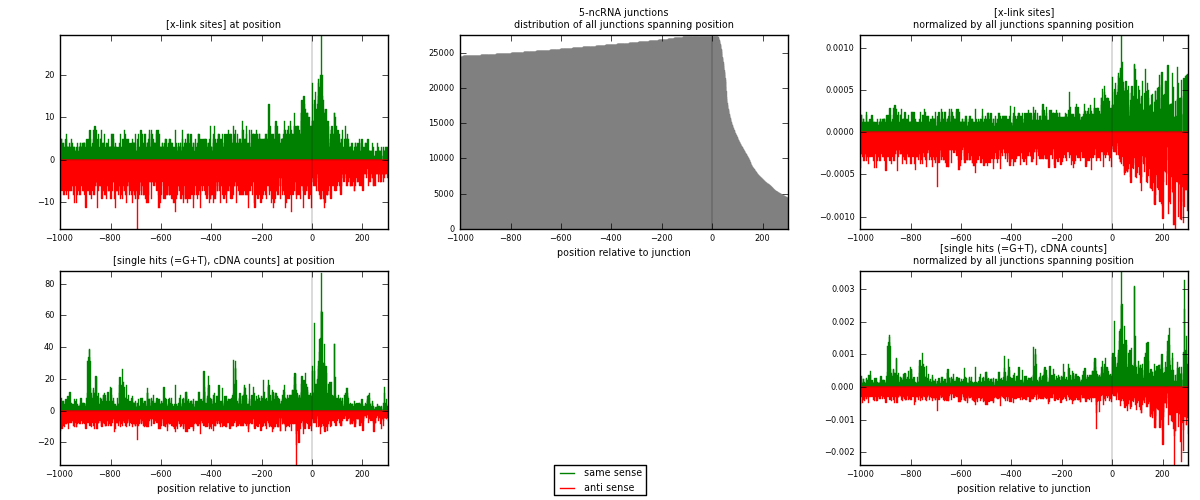 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4770 | 0.59% | 1.96% | 42.27% | 1847 | 1.18568 |
| anti | 6514 | 0.80% | 2.67% | 57.73% | 2292 | 1.61919 |
| both | 11284 | 1.39% | 4.63% | 100.00% | 4023 | 2.80487 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 8034 | 0.89% | 2.86% | 52.23% | 1847 | 1.99702 |
| anti | 7349 | 0.81% | 2.61% | 47.77% | 2292 | 1.82675 |
| both | 15383 | 1.70% | 5.47% | 100.00% | 4023 | 3.82376 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
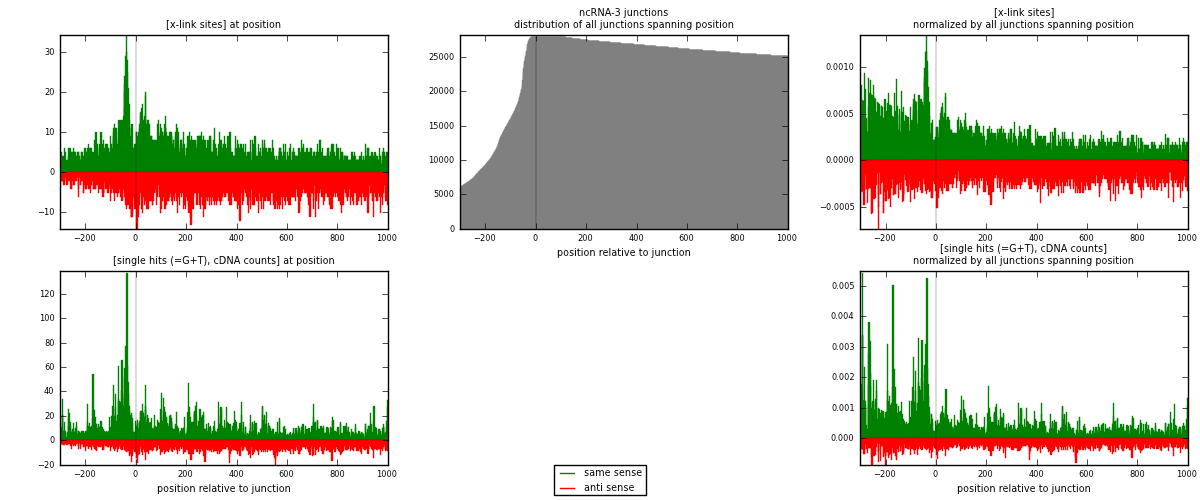 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6317 | 0.78% | 2.59% | 53.21% | 2188 | 1.53437 |
| anti | 5555 | 0.68% | 2.28% | 46.79% | 2032 | 1.34928 |
| both | 11872 | 1.46% | 4.87% | 100.00% | 4117 | 2.88365 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 11777 | 1.30% | 4.19% | 65.81% | 2188 | 2.86058 |
| anti | 6119 | 0.68% | 2.18% | 34.19% | 2032 | 1.48628 |
| both | 17896 | 1.98% | 6.36% | 100.00% | 4117 | 4.34685 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
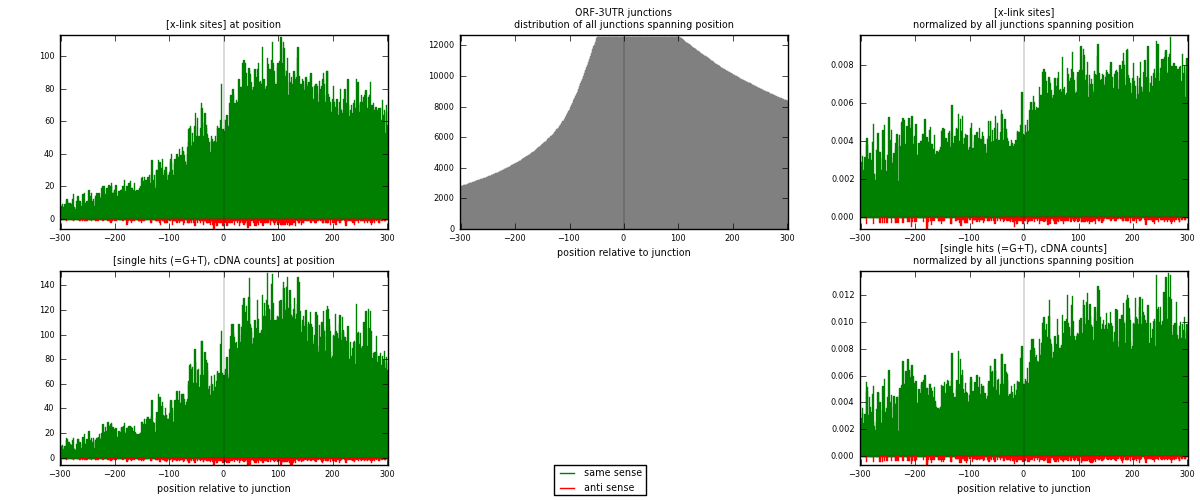
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 30321 | 3.74% | 12.44% | 98.25% | 5027 | 5.81419 |
| anti | 539 | 0.07% | 0.22% | 1.75% | 298 | 0.103356 |
| both | 30860 | 3.80% | 12.67% | 100.00% | 5215 | 5.91755 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 39330 | 4.35% | 13.99% | 98.56% | 5027 | 7.54171 |
| anti | 575 | 0.06% | 0.20% | 1.44% | 298 | 0.110259 |
| both | 39905 | 4.41% | 14.19% | 100.00% | 5215 | 7.65197 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
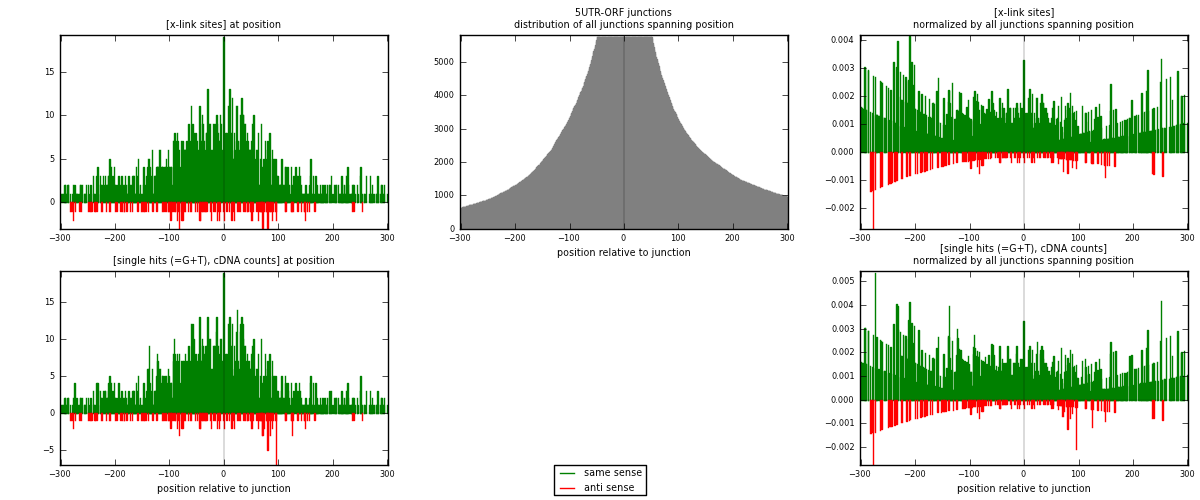
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1741 | 0.21% | 0.71% | 91.20% | 854 | 1.90899 |
| anti | 168 | 0.02% | 0.07% | 8.80% | 69 | 0.184211 |
| both | 1909 | 0.24% | 0.78% | 100.00% | 912 | 2.0932 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1853 | 0.20% | 0.66% | 90.97% | 854 | 2.0318 |
| anti | 184 | 0.02% | 0.07% | 9.03% | 69 | 0.201754 |
| both | 2037 | 0.23% | 0.72% | 100.00% | 912 | 2.23355 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
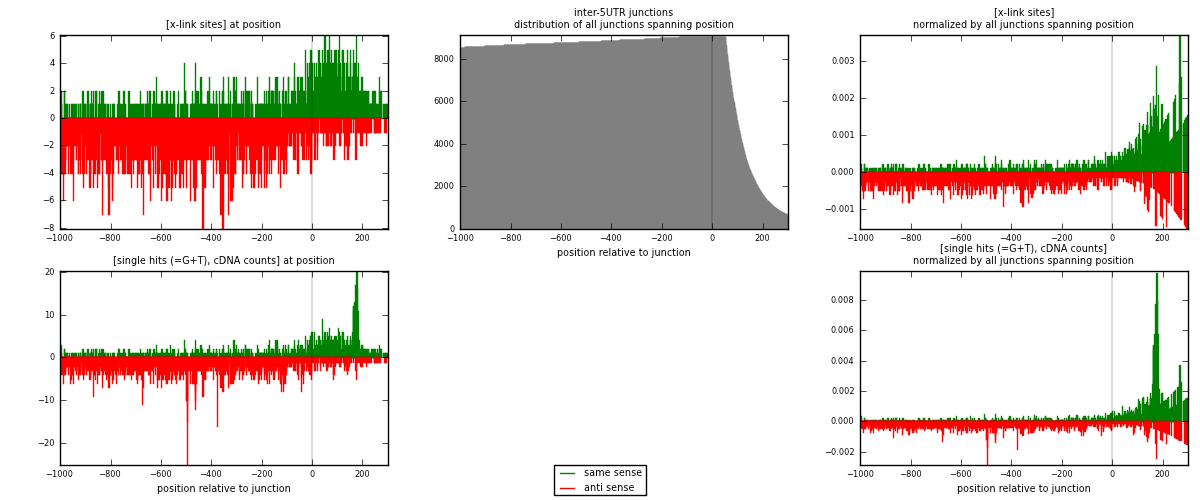
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1120 | 0.14% | 0.46% | 33.50% | 662 | 0.696951 |
| anti | 2223 | 0.27% | 0.91% | 66.50% | 1055 | 1.38332 |
| both | 3343 | 0.41% | 1.37% | 100.00% | 1607 | 2.08027 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1303 | 0.14% | 0.46% | 33.92% | 662 | 0.810828 |
| anti | 2538 | 0.28% | 0.90% | 66.08% | 1055 | 1.57934 |
| both | 3841 | 0.42% | 1.37% | 100.00% | 1607 | 2.39017 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
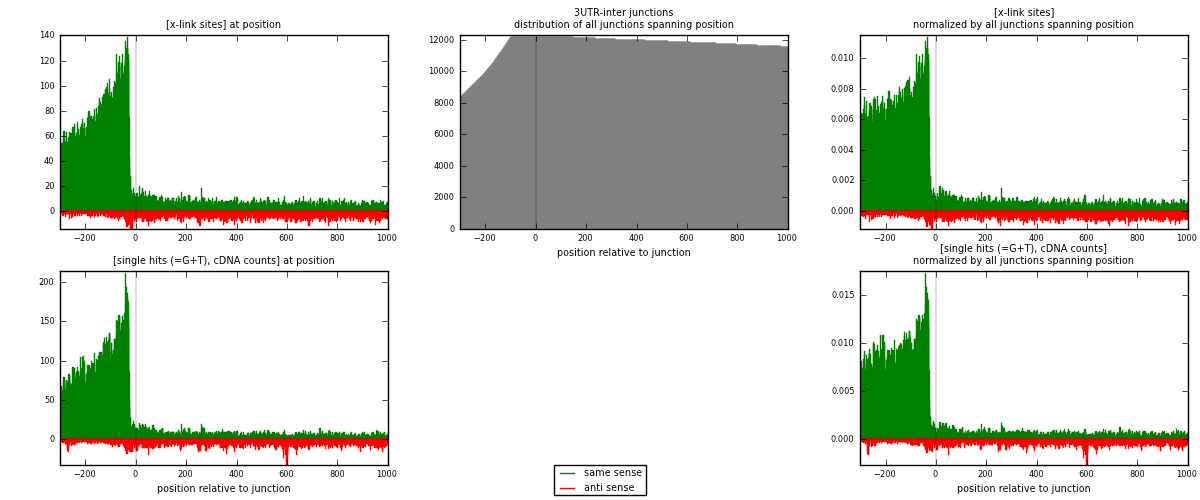
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 26764 | 3.30% | 10.98% | 84.87% | 4907 | 4.76992 |
| anti | 4773 | 0.59% | 1.96% | 15.13% | 1120 | 0.850651 |
| both | 31537 | 3.89% | 12.94% | 100.00% | 5611 | 5.62057 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 33085 | 3.66% | 11.76% | 84.63% | 4907 | 5.89645 |
| anti | 6007 | 0.66% | 2.14% | 15.37% | 1120 | 1.07058 |
| both | 39092 | 4.32% | 13.90% | 100.00% | 5611 | 6.96703 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
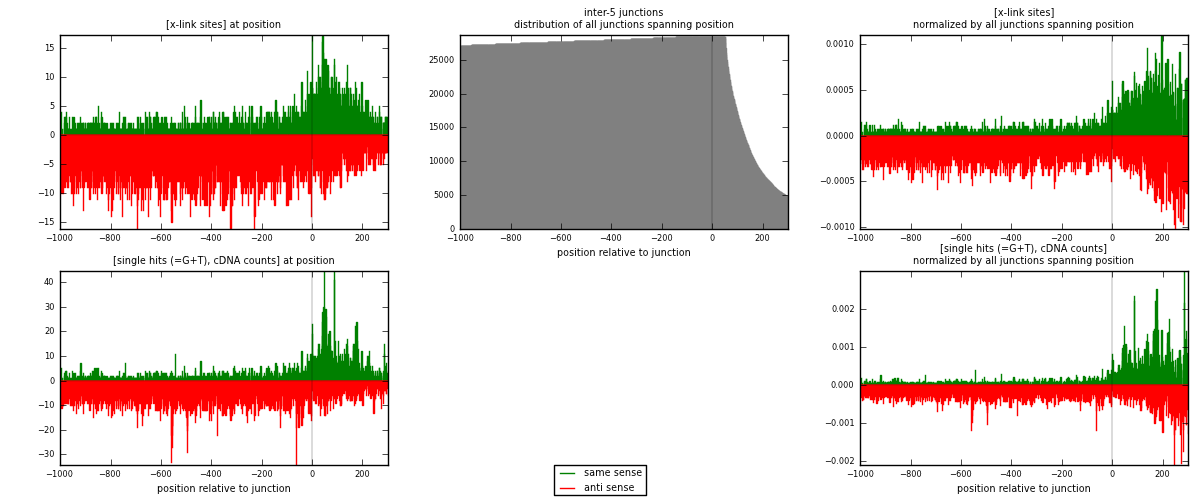
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2861 | 0.35% | 1.17% | 26.71% | 1448 | 0.67018 |
| anti | 7850 | 0.97% | 3.22% | 73.29% | 3024 | 1.83884 |
| both | 10711 | 1.32% | 4.40% | 100.00% | 4269 | 2.50902 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3765 | 0.42% | 1.34% | 29.34% | 1448 | 0.88194 |
| anti | 9066 | 1.00% | 3.22% | 70.66% | 3024 | 2.12368 |
| both | 12831 | 1.42% | 4.56% | 100.00% | 4269 | 3.00562 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
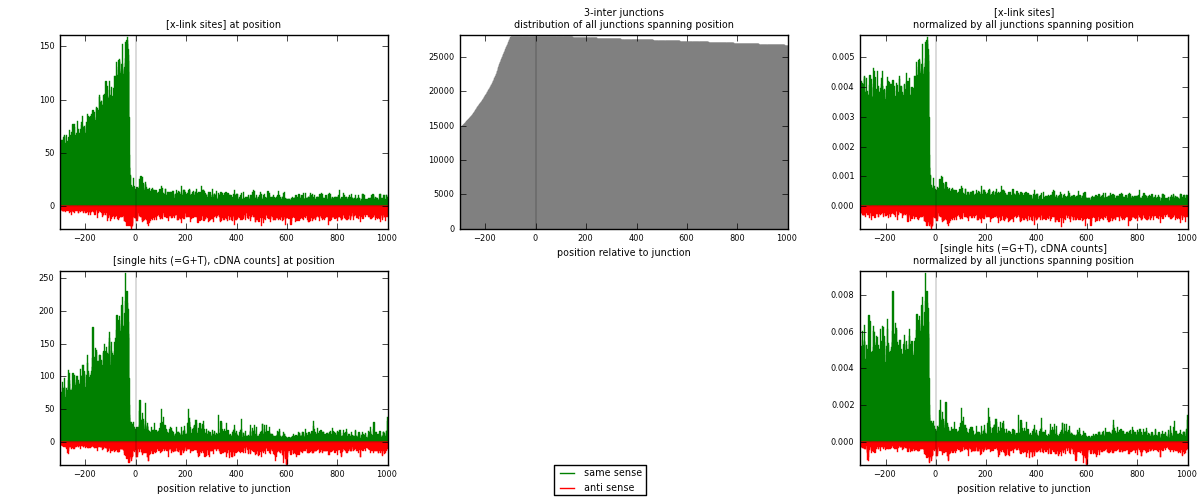
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 32223 | 3.97% | 13.23% | 77.39% | 6273 | 3.92198 |
| anti | 9415 | 1.16% | 3.86% | 22.61% | 2486 | 1.14593 |
| both | 41638 | 5.13% | 17.09% | 100.00% | 8216 | 5.06792 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 44118 | 4.88% | 15.69% | 79.42% | 6273 | 5.36977 |
| anti | 11432 | 1.26% | 4.07% | 20.58% | 2486 | 1.39143 |
| both | 55550 | 6.14% | 19.75% | 100.00% | 8216 | 6.7612 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
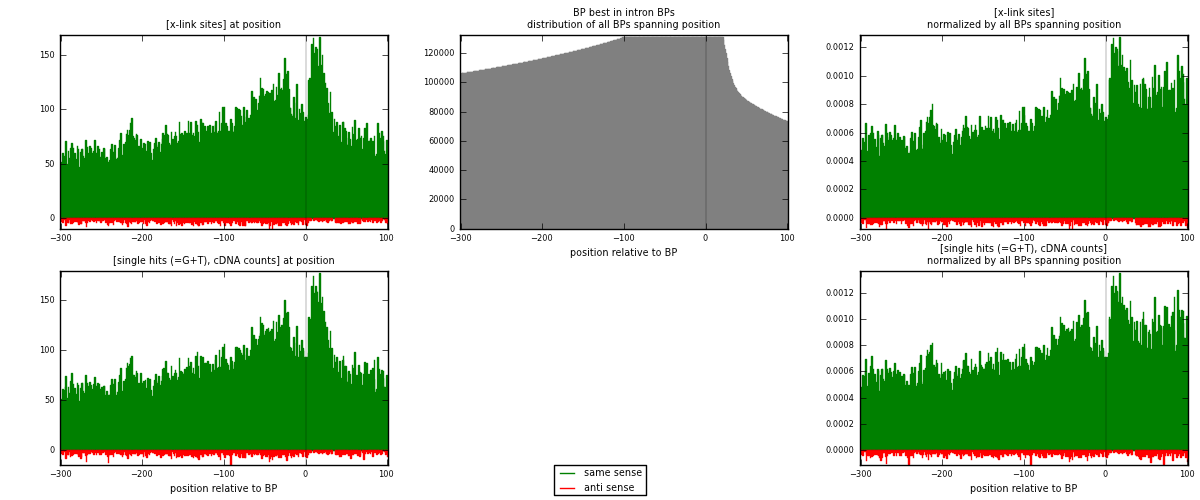
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 32683 | 4.03% | 13.41% | 96.33% | 20928 | 1.5026 |
| anti | 1246 | 0.15% | 0.51% | 3.67% | 962 | 0.0572847 |
| both | 33929 | 4.18% | 13.93% | 100.00% | 21751 | 1.55988 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 33905 | 3.75% | 12.06% | 96.20% | 20928 | 1.55878 |
| anti | 1341 | 0.15% | 0.48% | 3.80% | 962 | 0.0616523 |
| both | 35246 | 3.90% | 12.53% | 100.00% | 21751 | 1.62043 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.