RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
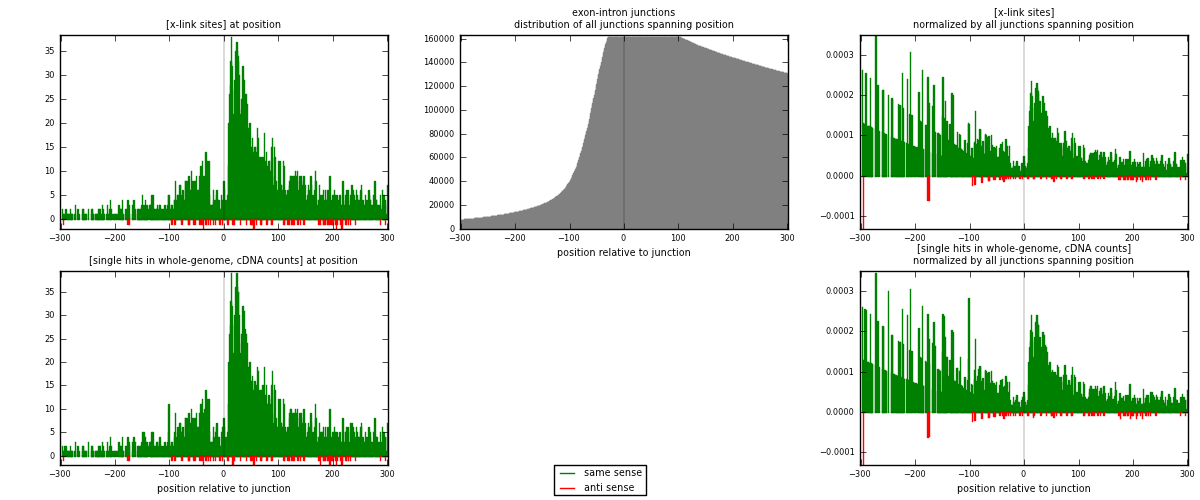 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3272 | 9.51% | 29.27% | 97.67% | 2989 | 1.06928 |
| anti | 78 | 0.23% | 0.70% | 2.33% | 71 | 0.0254902 |
| both | 3350 | 9.74% | 29.97% | 100.00% | 3060 | 1.09477 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3367 | 9.31% | 28.39% | 97.65% | 2989 | 1.10033 |
| anti | 81 | 0.22% | 0.68% | 2.35% | 71 | 0.0264706 |
| both | 3448 | 9.53% | 29.07% | 100.00% | 3060 | 1.1268 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
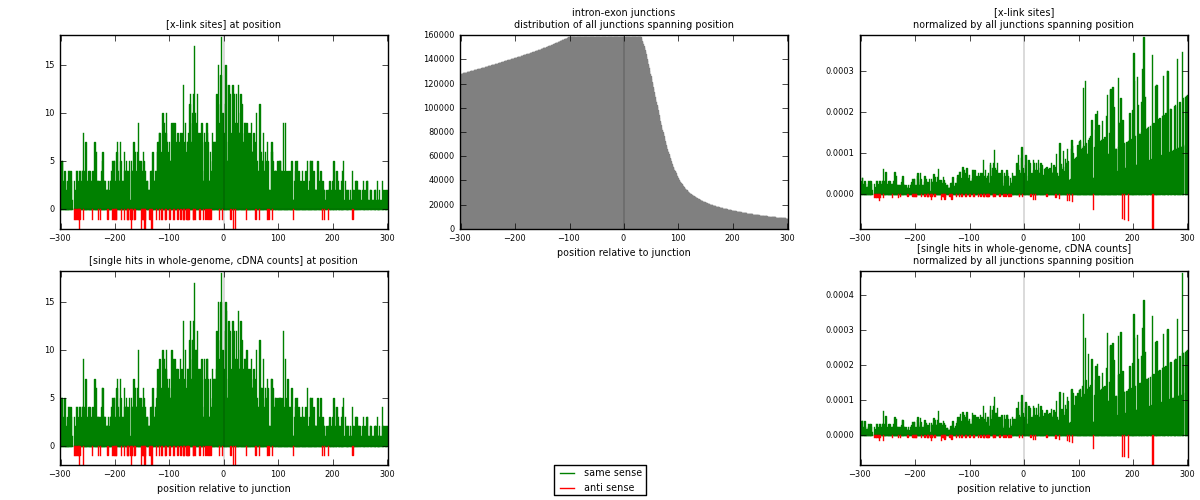 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2390 | 6.95% | 21.38% | 96.53% | 2134 | 1.08096 |
| anti | 86 | 0.25% | 0.77% | 3.47% | 77 | 0.0388964 |
| both | 2476 | 7.20% | 22.15% | 100.00% | 2211 | 1.11986 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2460 | 6.80% | 20.74% | 96.58% | 2134 | 1.11262 |
| anti | 87 | 0.24% | 0.73% | 3.42% | 77 | 0.0393487 |
| both | 2547 | 7.04% | 21.48% | 100.00% | 2211 | 1.15197 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
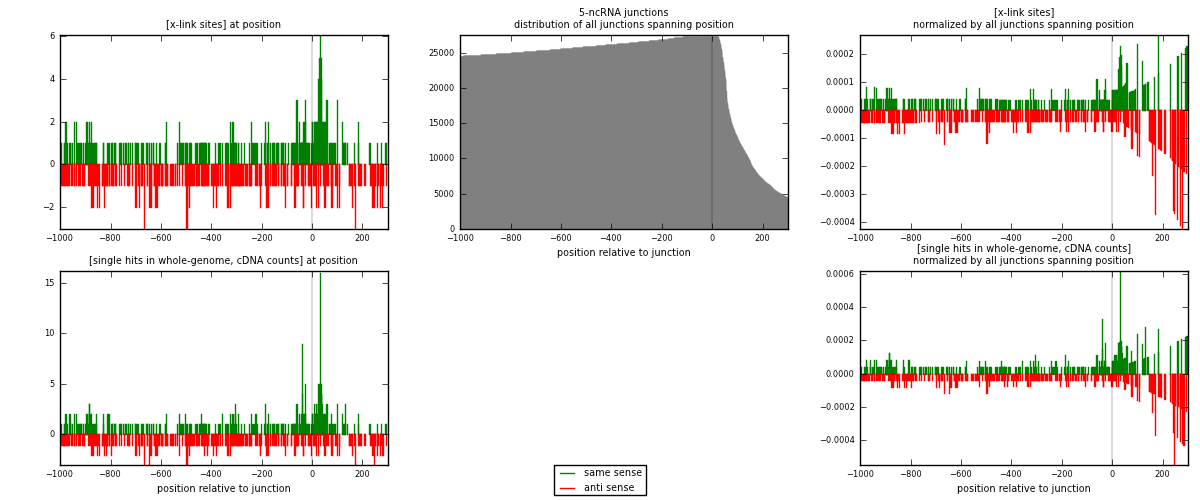 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 332 | 0.96% | 2.97% | 49.63% | 181 | 0.772093 |
| anti | 337 | 0.98% | 3.02% | 50.37% | 250 | 0.783721 |
| both | 669 | 1.94% | 5.99% | 100.00% | 430 | 1.55581 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 392 | 1.08% | 3.31% | 53.19% | 181 | 0.911628 |
| anti | 345 | 0.95% | 2.91% | 46.81% | 250 | 0.802326 |
| both | 737 | 2.04% | 6.21% | 100.00% | 430 | 1.71395 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
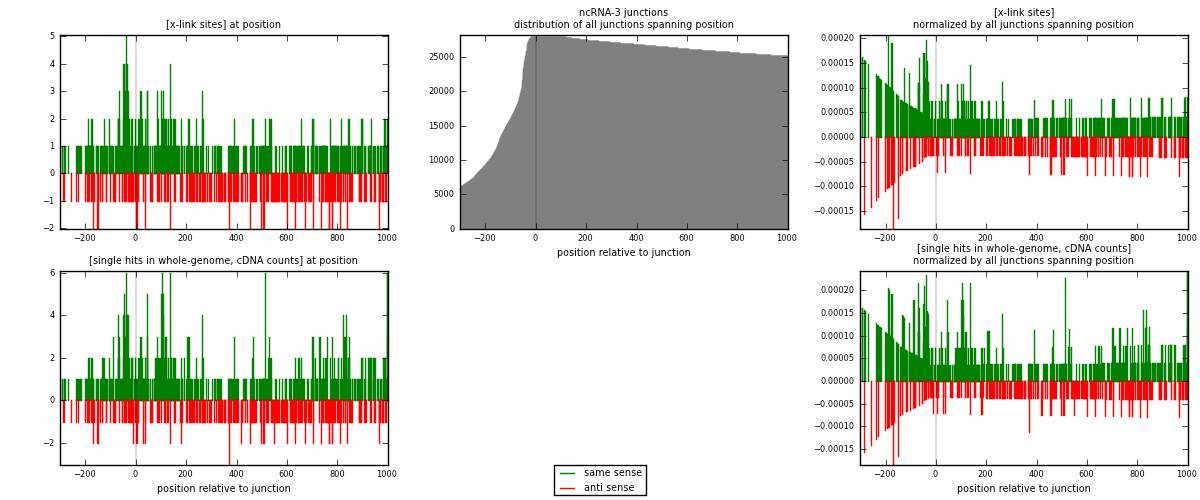 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 495 | 1.44% | 4.43% | 65.56% | 263 | 1.06911 |
| anti | 260 | 0.76% | 2.33% | 34.44% | 201 | 0.561555 |
| both | 755 | 2.19% | 6.75% | 100.00% | 463 | 1.63067 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 596 | 1.65% | 5.03% | 69.14% | 263 | 1.28726 |
| anti | 266 | 0.74% | 2.24% | 30.86% | 201 | 0.574514 |
| both | 862 | 2.38% | 7.27% | 100.00% | 463 | 1.86177 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
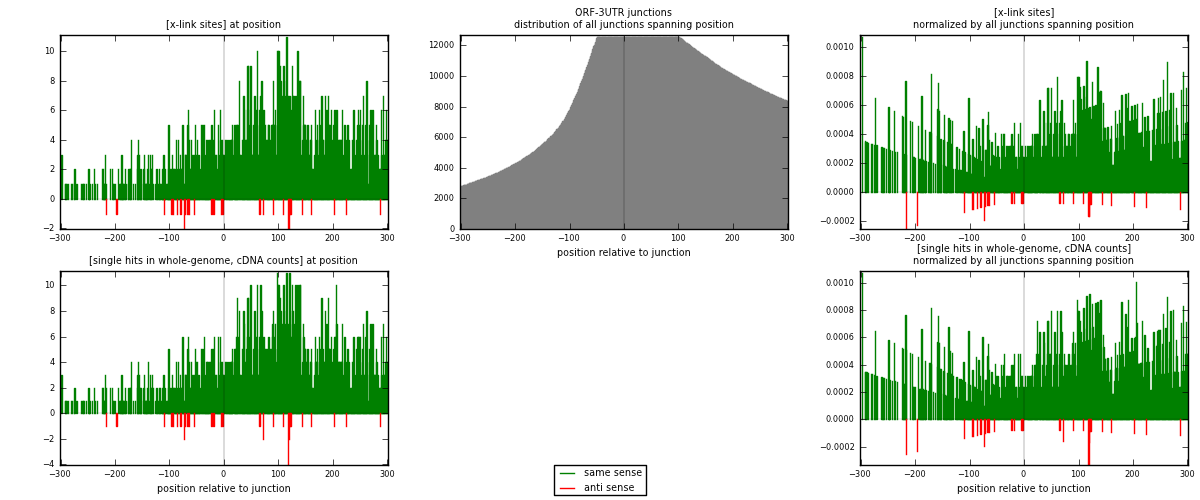 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1597 | 4.64% | 14.29% | 98.04% | 928 | 1.67576 |
| anti | 32 | 0.09% | 0.29% | 1.96% | 25 | 0.0335782 |
| both | 1629 | 4.73% | 14.57% | 100.00% | 953 | 1.70934 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1705 | 4.71% | 14.38% | 97.99% | 928 | 1.78909 |
| anti | 35 | 0.10% | 0.30% | 2.01% | 25 | 0.0367261 |
| both | 1740 | 4.81% | 14.67% | 100.00% | 953 | 1.82581 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
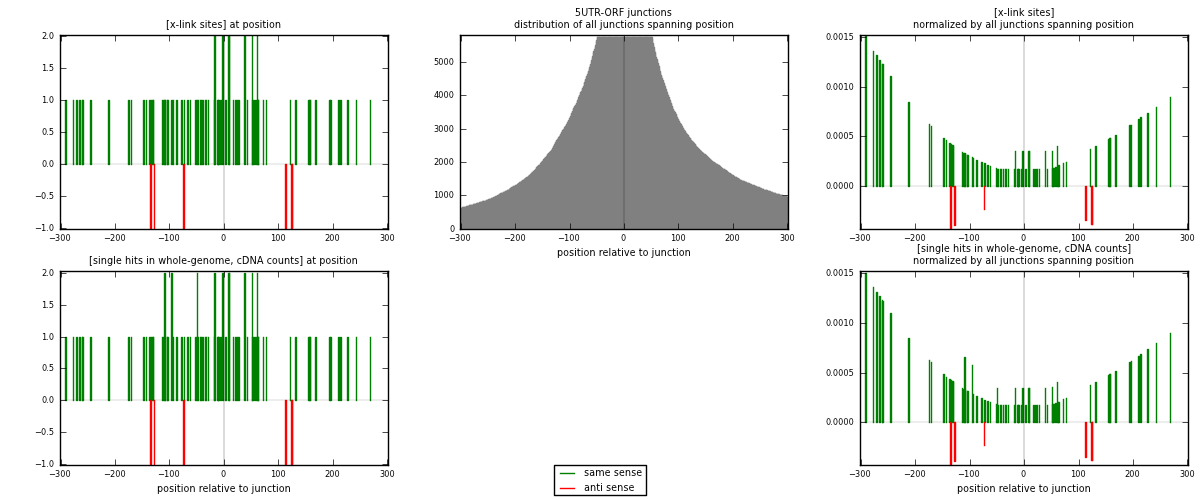 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 83 | 0.24% | 0.74% | 93.26% | 75 | 1.0641 |
| anti | 6 | 0.02% | 0.05% | 6.74% | 3 | 0.0769231 |
| both | 89 | 0.26% | 0.80% | 100.00% | 78 | 1.14103 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 86 | 0.24% | 0.73% | 93.48% | 75 | 1.10256 |
| anti | 6 | 0.02% | 0.05% | 6.52% | 3 | 0.0769231 |
| both | 92 | 0.25% | 0.78% | 100.00% | 78 | 1.17949 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
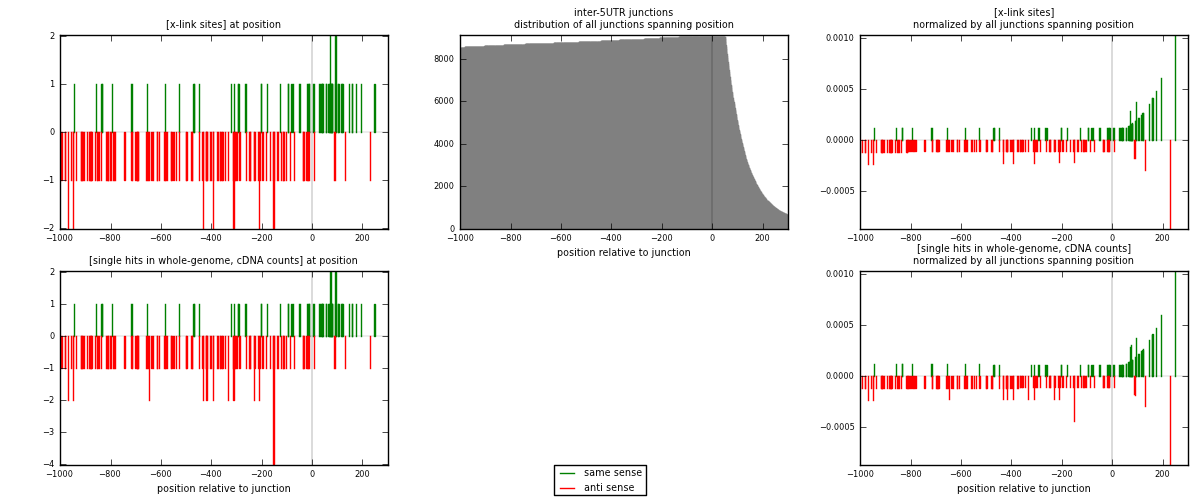 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 68 | 0.20% | 0.61% | 33.83% | 65 | 0.412121 |
| anti | 133 | 0.39% | 1.19% | 66.17% | 100 | 0.806061 |
| both | 201 | 0.58% | 1.80% | 100.00% | 165 | 1.21818 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 69 | 0.19% | 0.58% | 33.17% | 65 | 0.418182 |
| anti | 139 | 0.38% | 1.17% | 66.83% | 100 | 0.842424 |
| both | 208 | 0.57% | 1.75% | 100.00% | 165 | 1.26061 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
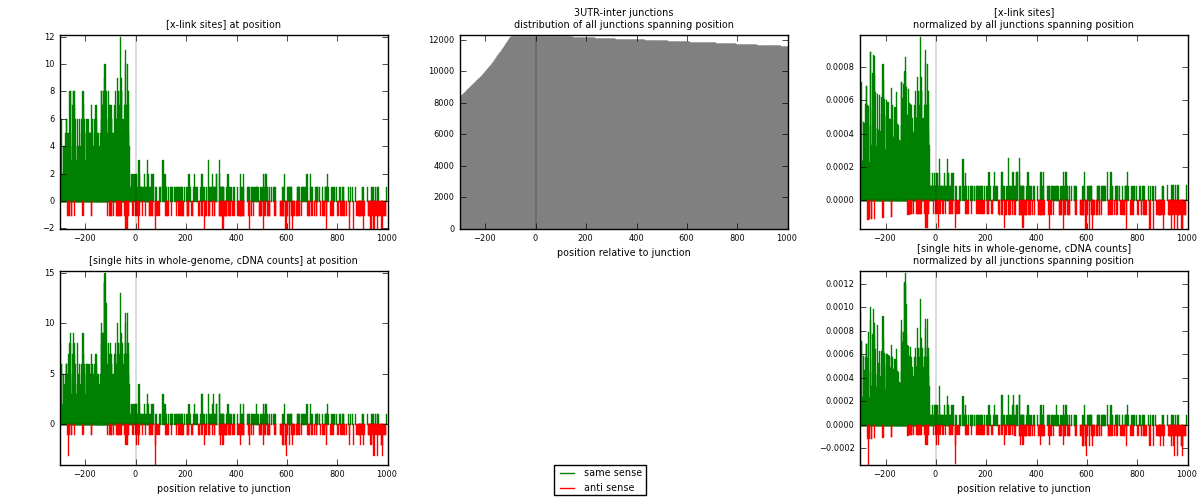 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1377 | 4.00% | 12.32% | 84.07% | 860 | 1.36067 |
| anti | 261 | 0.76% | 2.34% | 15.93% | 162 | 0.257905 |
| both | 1638 | 4.76% | 14.66% | 100.00% | 1012 | 1.61858 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1480 | 4.09% | 12.48% | 84.09% | 860 | 1.46245 |
| anti | 280 | 0.77% | 2.36% | 15.91% | 162 | 0.27668 |
| both | 1760 | 4.86% | 14.84% | 100.00% | 1012 | 1.73913 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
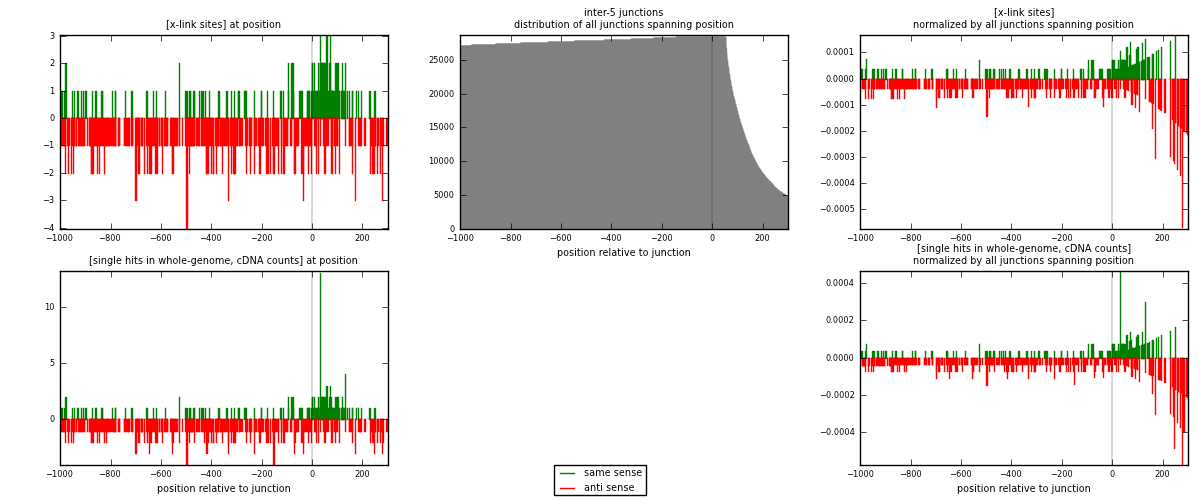
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 175 | 0.51% | 1.57% | 28.18% | 142 | 0.380435 |
| anti | 446 | 1.30% | 3.99% | 71.82% | 318 | 0.969565 |
| both | 621 | 1.80% | 5.56% | 100.00% | 460 | 1.35 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 188 | 0.52% | 1.59% | 28.88% | 142 | 0.408696 |
| anti | 463 | 1.28% | 3.90% | 71.12% | 318 | 1.00652 |
| both | 651 | 1.80% | 5.49% | 100.00% | 460 | 1.41522 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
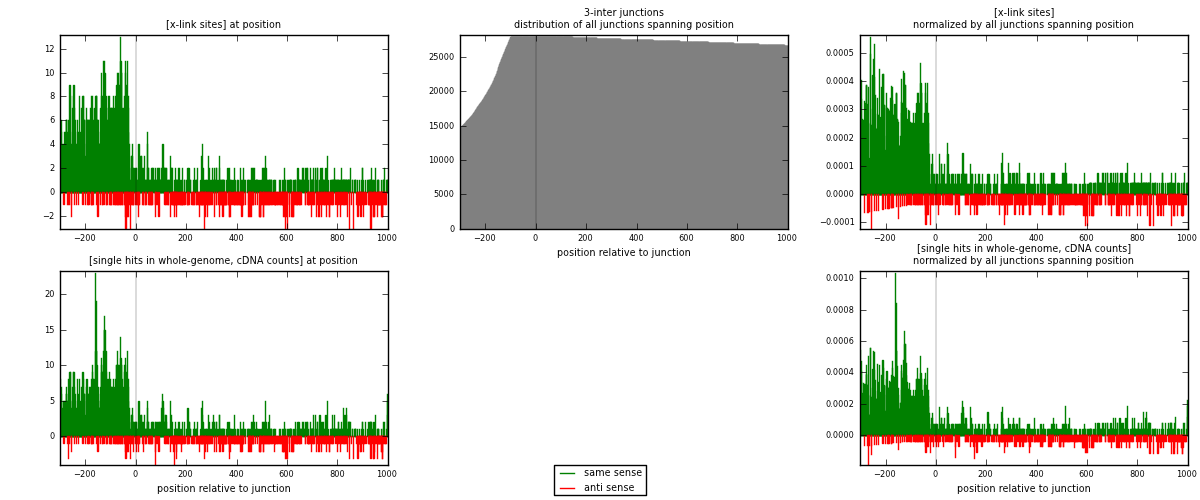
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1846 | 5.37% | 16.52% | 78.59% | 1066 | 1.35139 |
| anti | 503 | 1.46% | 4.50% | 21.41% | 311 | 0.368228 |
| both | 2349 | 6.83% | 21.02% | 100.00% | 1366 | 1.71962 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2112 | 5.84% | 17.81% | 79.79% | 1066 | 1.54612 |
| anti | 535 | 1.48% | 4.51% | 20.21% | 311 | 0.391654 |
| both | 2647 | 7.32% | 22.32% | 100.00% | 1366 | 1.93777 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
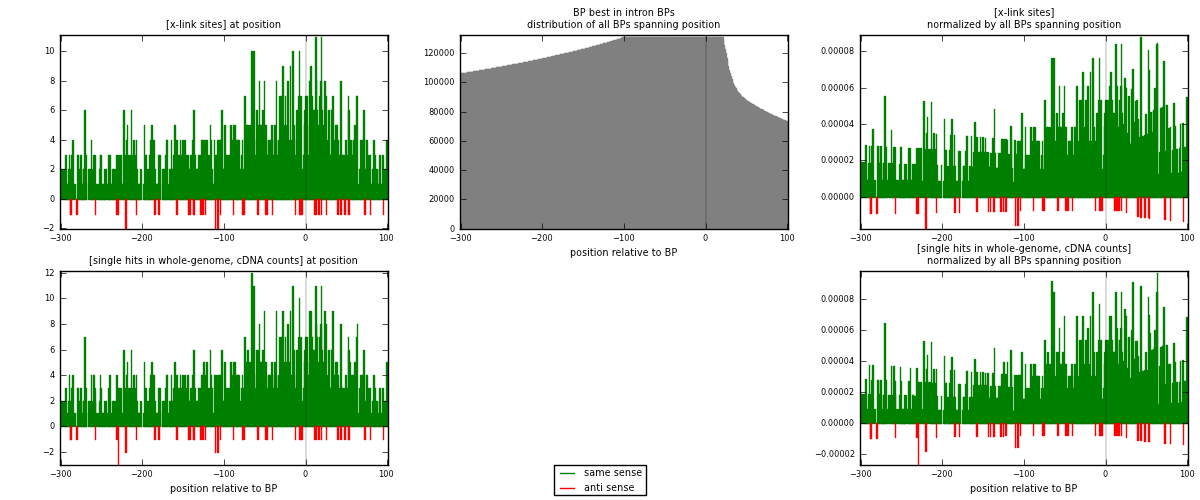
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 1268 | 3.69% | 11.34% | 96.35% | 1195 | 1.02258 |
| anti | 48 | 0.14% | 0.43% | 3.65% | 45 | 0.0387097 |
| both | 1316 | 3.82% | 11.77% | 100.00% | 1240 | 1.06129 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 1307 | 3.61% | 11.02% | 96.32% | 1195 | 1.05403 |
| anti | 50 | 0.14% | 0.42% | 3.68% | 45 | 0.0403226 |
| both | 1357 | 3.75% | 11.44% | 100.00% | 1240 | 1.09435 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.