RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
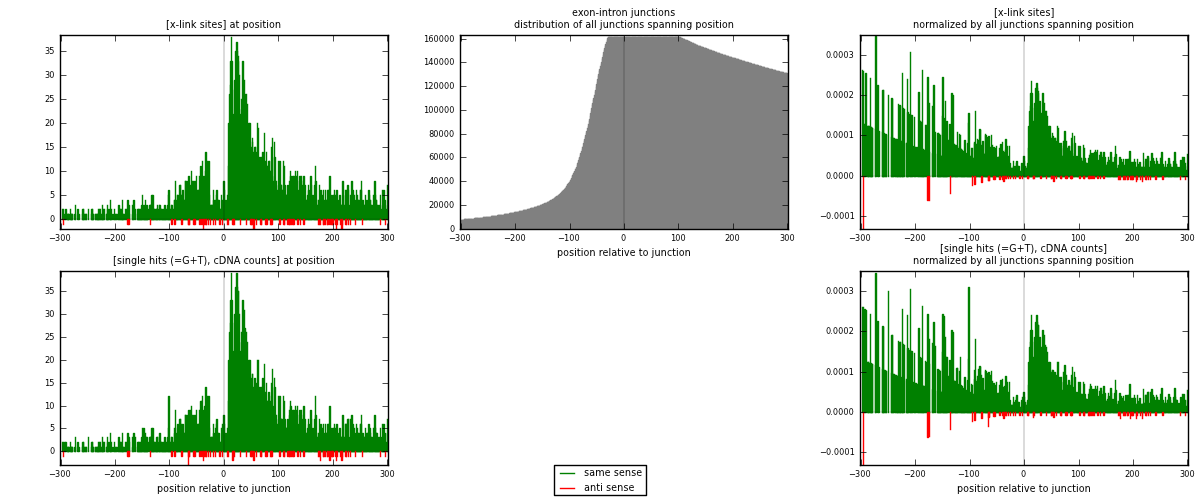 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3332 | 9.19% | 28.67% | 97.51% | 3041 | 1.06863 |
| anti | 85 | 0.23% | 0.73% | 2.49% | 77 | 0.0272611 |
| both | 3417 | 9.43% | 29.41% | 100.00% | 3118 | 1.09589 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3430 | 8.96% | 27.72% | 97.44% | 3041 | 1.10006 |
| anti | 90 | 0.23% | 0.73% | 2.56% | 77 | 0.0288647 |
| both | 3520 | 9.19% | 28.44% | 100.00% | 3118 | 1.12893 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
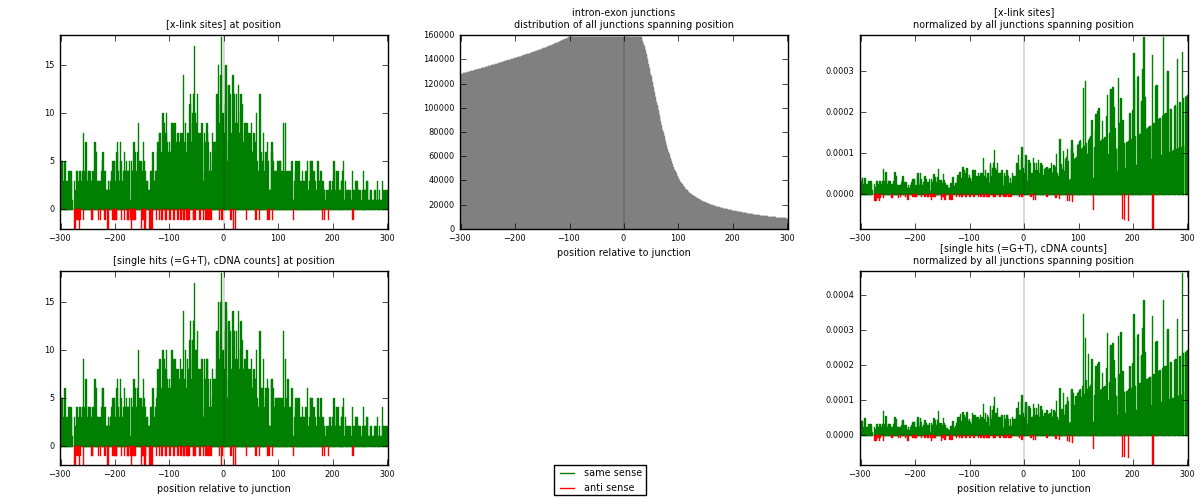 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2435 | 6.72% | 20.96% | 96.28% | 2167 | 1.08174 |
| anti | 94 | 0.26% | 0.81% | 3.72% | 84 | 0.0417592 |
| both | 2529 | 6.98% | 21.76% | 100.00% | 2251 | 1.1235 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2506 | 6.54% | 20.25% | 96.31% | 2167 | 1.11328 |
| anti | 96 | 0.25% | 0.78% | 3.69% | 84 | 0.0426477 |
| both | 2602 | 6.79% | 21.03% | 100.00% | 2251 | 1.15593 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
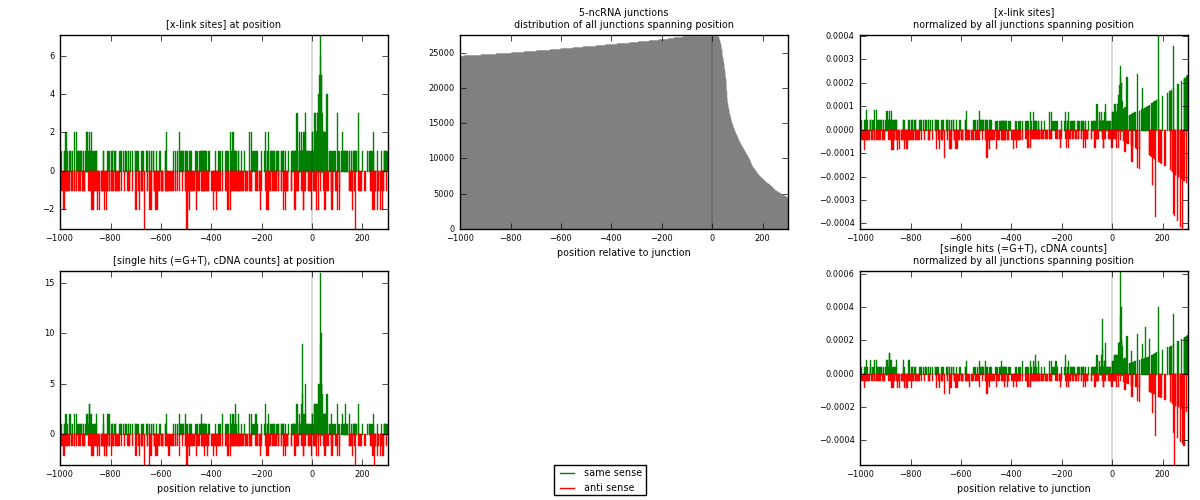 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 379 | 1.05% | 3.26% | 51.49% | 199 | 0.823913 |
| anti | 357 | 0.98% | 3.07% | 48.51% | 262 | 0.776087 |
| both | 736 | 2.03% | 6.33% | 100.00% | 460 | 1.6 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 447 | 1.17% | 3.61% | 54.98% | 199 | 0.971739 |
| anti | 366 | 0.96% | 2.96% | 45.02% | 262 | 0.795652 |
| both | 813 | 2.12% | 6.57% | 100.00% | 460 | 1.76739 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
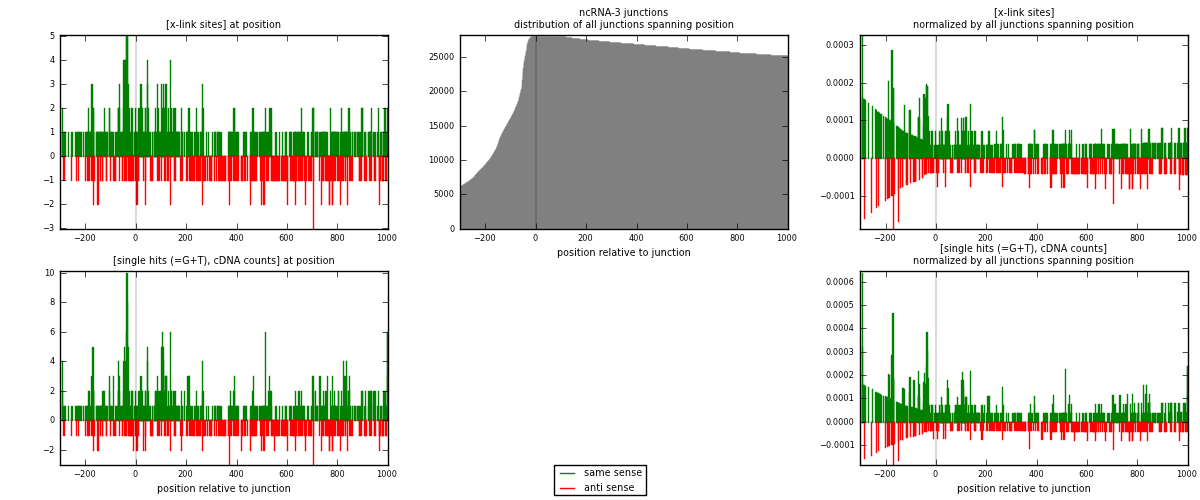 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 545 | 1.50% | 4.69% | 66.79% | 283 | 1.11224 |
| anti | 271 | 0.75% | 2.33% | 33.21% | 208 | 0.553061 |
| both | 816 | 2.25% | 7.02% | 100.00% | 490 | 1.66531 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 665 | 1.74% | 5.37% | 70.59% | 283 | 1.35714 |
| anti | 277 | 0.72% | 2.24% | 29.41% | 208 | 0.565306 |
| both | 942 | 2.46% | 7.61% | 100.00% | 490 | 1.92245 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
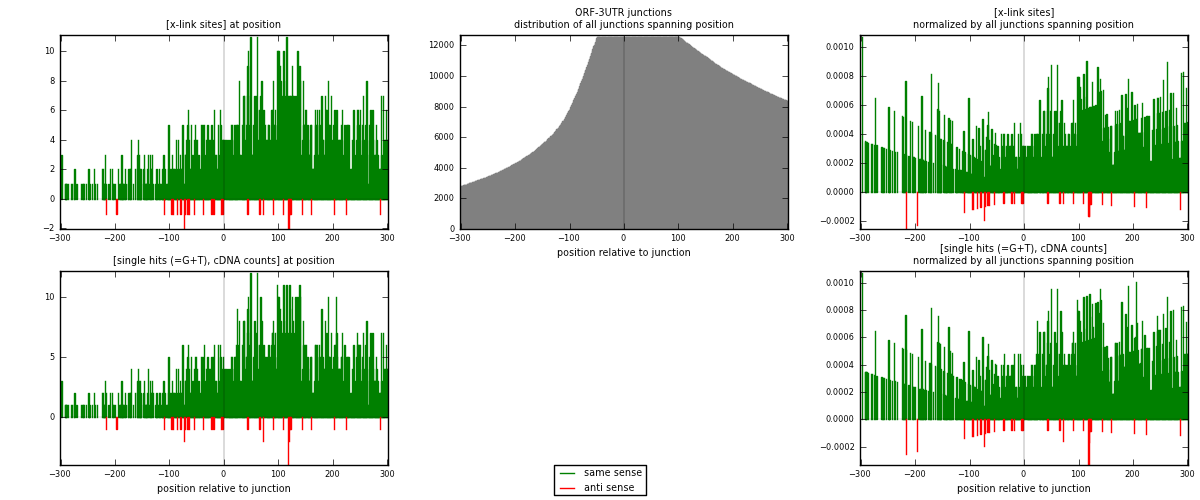
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1680 | 4.63% | 14.46% | 98.02% | 957 | 1.70732 |
| anti | 34 | 0.09% | 0.29% | 1.98% | 27 | 0.0345528 |
| both | 1714 | 4.73% | 14.75% | 100.00% | 984 | 1.74187 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1793 | 4.68% | 14.49% | 97.98% | 957 | 1.82215 |
| anti | 37 | 0.10% | 0.30% | 2.02% | 27 | 0.0376016 |
| both | 1830 | 4.78% | 14.79% | 100.00% | 984 | 1.85976 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
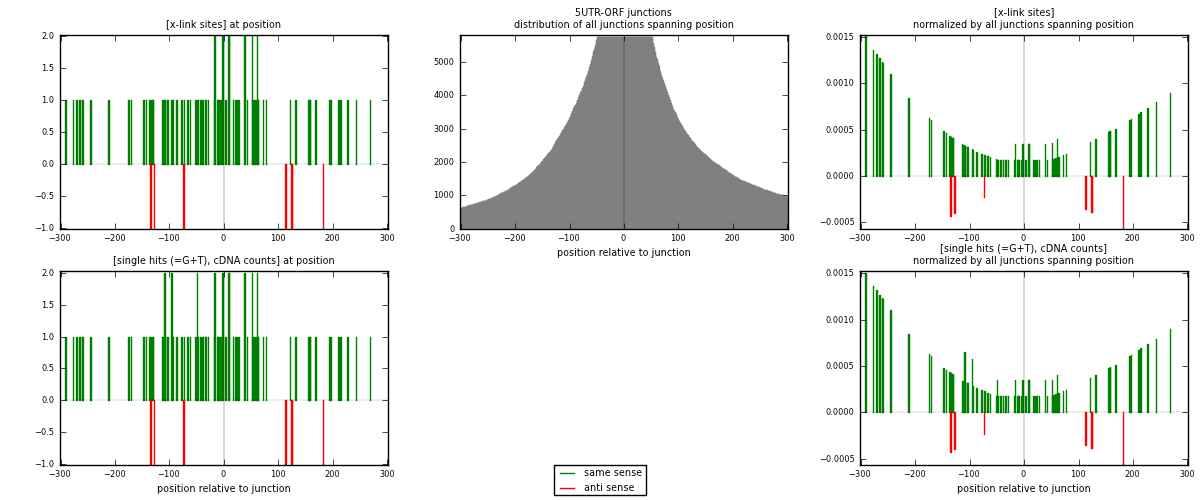
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 83 | 0.23% | 0.71% | 92.22% | 75 | 1.05063 |
| anti | 7 | 0.02% | 0.06% | 7.78% | 4 | 0.0886076 |
| both | 90 | 0.25% | 0.77% | 100.00% | 79 | 1.13924 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 86 | 0.22% | 0.69% | 92.47% | 75 | 1.08861 |
| anti | 7 | 0.02% | 0.06% | 7.53% | 4 | 0.0886076 |
| both | 93 | 0.24% | 0.75% | 100.00% | 79 | 1.17722 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
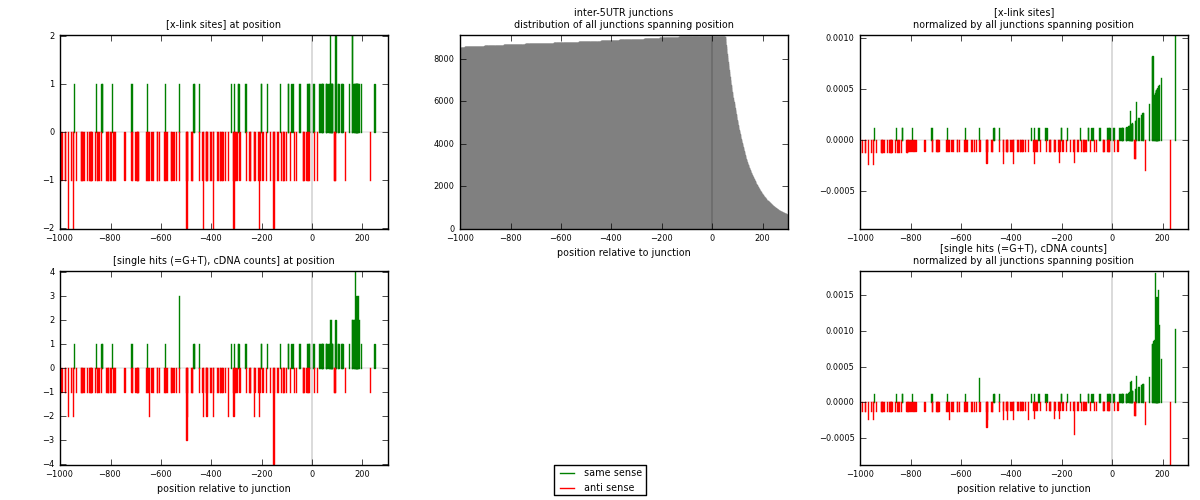
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 83 | 0.23% | 0.71% | 37.39% | 67 | 0.488235 |
| anti | 139 | 0.38% | 1.20% | 62.61% | 103 | 0.817647 |
| both | 222 | 0.61% | 1.91% | 100.00% | 170 | 1.30588 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 97 | 0.25% | 0.78% | 39.75% | 67 | 0.570588 |
| anti | 147 | 0.38% | 1.19% | 60.25% | 103 | 0.864706 |
| both | 244 | 0.64% | 1.97% | 100.00% | 170 | 1.43529 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
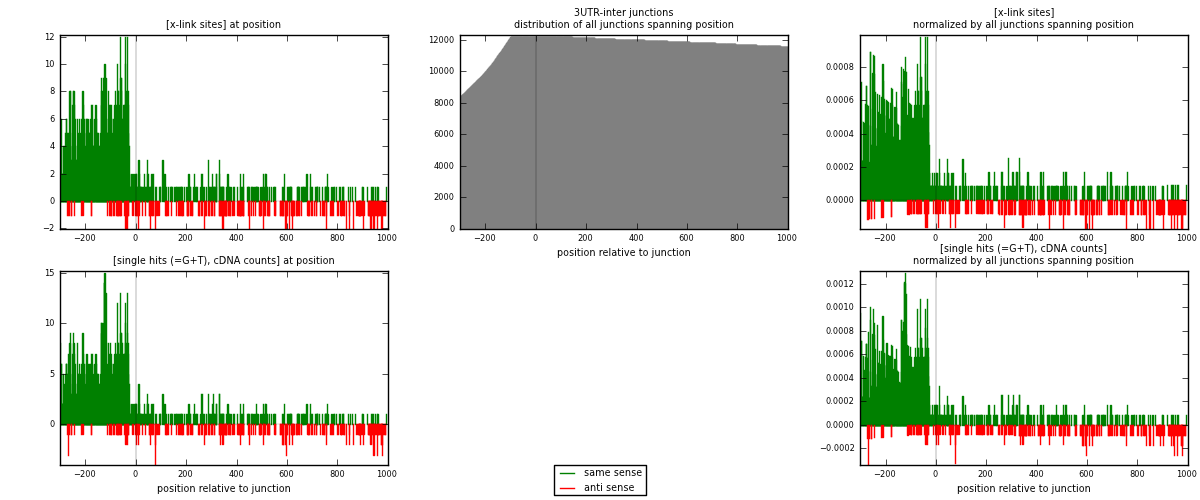
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1435 | 3.96% | 12.35% | 83.97% | 890 | 1.36407 |
| anti | 274 | 0.76% | 2.36% | 16.03% | 172 | 0.260456 |
| both | 1709 | 4.71% | 14.71% | 100.00% | 1052 | 1.62452 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1540 | 4.02% | 12.44% | 84.02% | 890 | 1.46388 |
| anti | 293 | 0.77% | 2.37% | 15.98% | 172 | 0.278517 |
| both | 1833 | 4.79% | 14.81% | 100.00% | 1052 | 1.7424 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
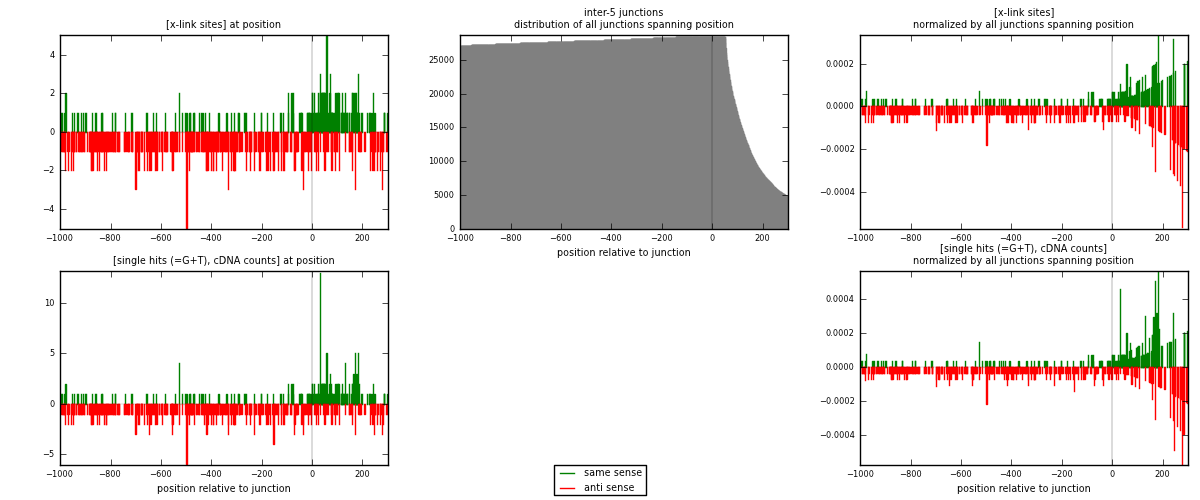
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 220 | 0.61% | 1.89% | 32.26% | 149 | 0.463158 |
| anti | 462 | 1.27% | 3.98% | 67.74% | 326 | 0.972632 |
| both | 682 | 1.88% | 5.87% | 100.00% | 475 | 1.43579 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 247 | 0.64% | 2.00% | 33.93% | 149 | 0.52 |
| anti | 481 | 1.26% | 3.89% | 66.07% | 326 | 1.01263 |
| both | 728 | 1.90% | 5.88% | 100.00% | 475 | 1.53263 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
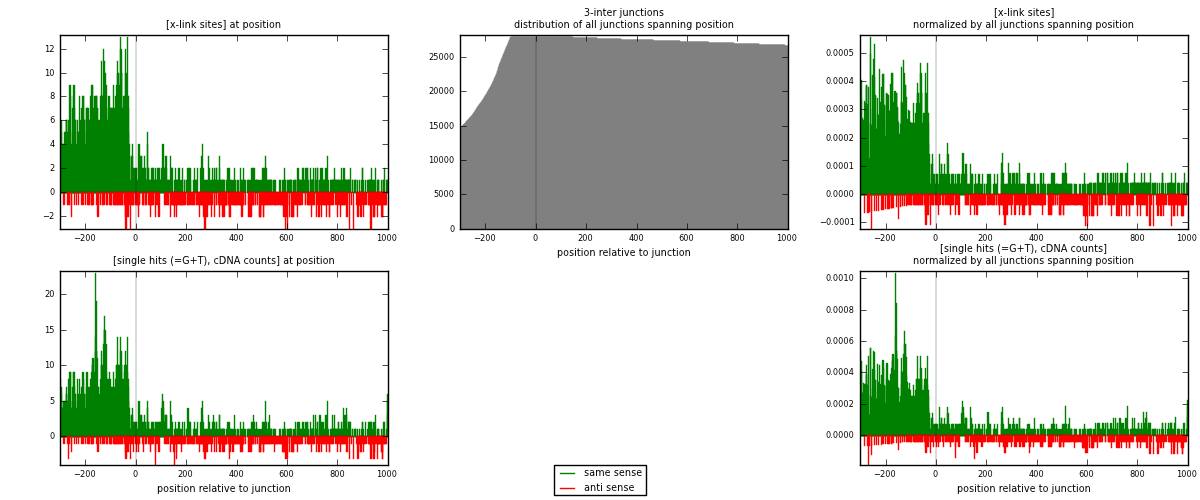
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1941 | 5.35% | 16.70% | 78.68% | 1105 | 1.36883 |
| anti | 526 | 1.45% | 4.53% | 21.32% | 325 | 0.370945 |
| both | 2467 | 6.81% | 21.23% | 100.00% | 1418 | 1.73977 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2238 | 5.84% | 18.08% | 80.01% | 1105 | 1.57828 |
| anti | 559 | 1.46% | 4.52% | 19.99% | 325 | 0.394217 |
| both | 2797 | 7.30% | 22.60% | 100.00% | 1418 | 1.9725 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
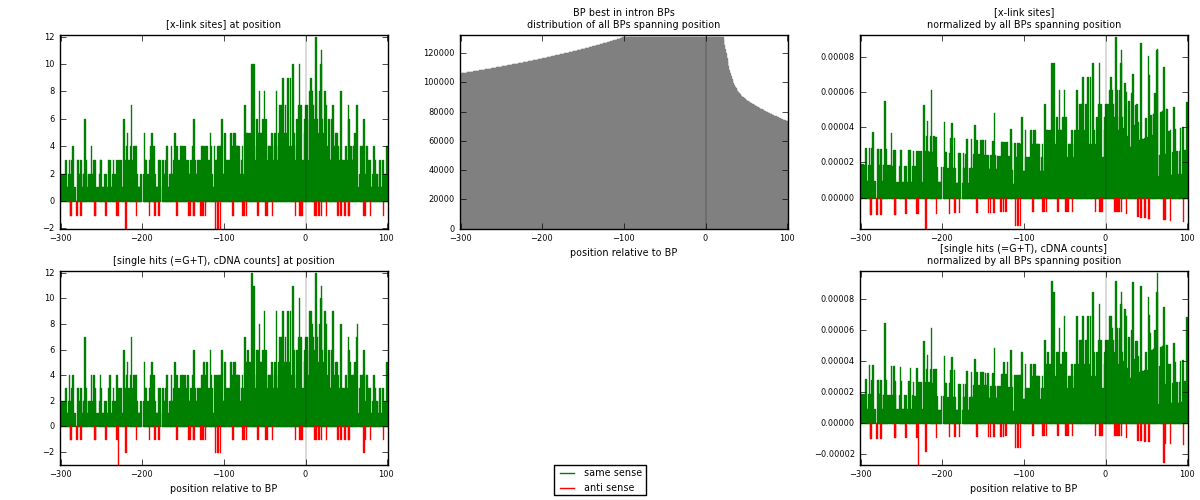
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 1296 | 3.57% | 11.15% | 95.86% | 1221 | 1.01727 |
| anti | 56 | 0.15% | 0.48% | 4.14% | 53 | 0.043956 |
| both | 1352 | 3.73% | 11.64% | 100.00% | 1274 | 1.06122 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 1335 | 3.49% | 10.79% | 95.77% | 1221 | 1.04788 |
| anti | 59 | 0.15% | 0.48% | 4.23% | 53 | 0.0463108 |
| both | 1394 | 3.64% | 11.26% | 100.00% | 1274 | 1.09419 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.