RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
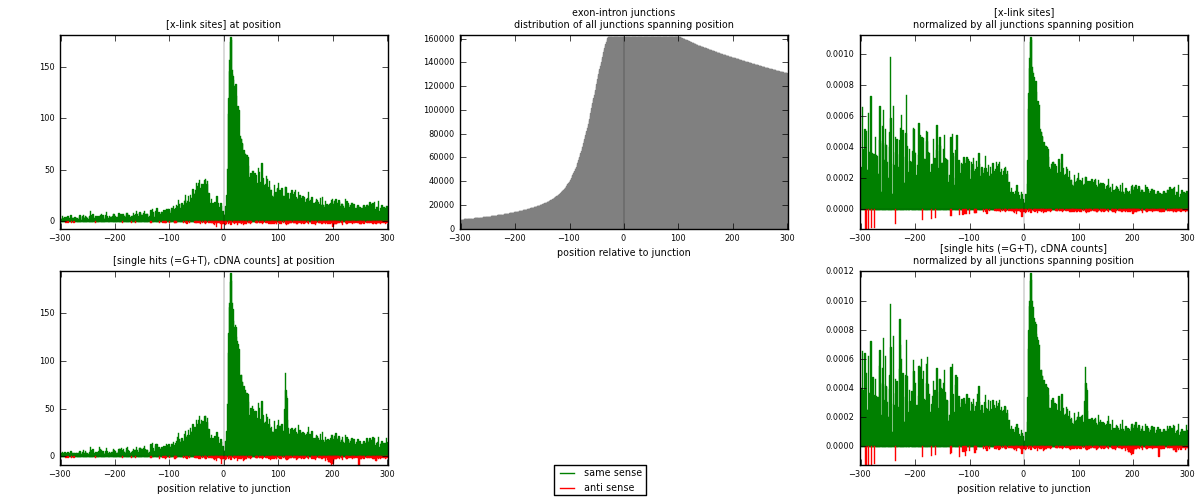 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12370 | 8.96% | 27.84% | 97.41% | 9946 | 1.21824 |
| anti | 329 | 0.24% | 0.74% | 2.59% | 235 | 0.032401 |
| both | 12699 | 9.20% | 28.58% | 100.00% | 10154 | 1.25064 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 13204 | 8.50% | 25.48% | 97.10% | 9946 | 1.30037 |
| anti | 394 | 0.25% | 0.76% | 2.90% | 235 | 0.0388024 |
| both | 13598 | 8.75% | 26.24% | 100.00% | 10154 | 1.33918 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
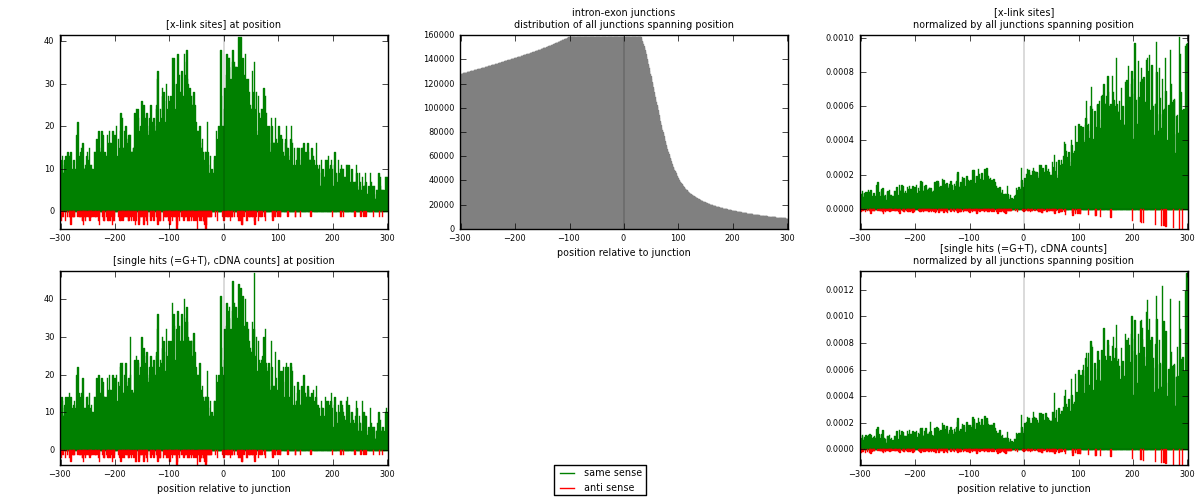 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9306 | 6.74% | 20.94% | 96.44% | 7299 | 1.22965 |
| anti | 344 | 0.25% | 0.77% | 3.56% | 282 | 0.0454545 |
| both | 9650 | 6.99% | 21.72% | 100.00% | 7568 | 1.27511 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 9942 | 6.40% | 19.18% | 96.58% | 7299 | 1.31369 |
| anti | 352 | 0.23% | 0.68% | 3.42% | 282 | 0.0465116 |
| both | 10294 | 6.62% | 19.86% | 100.00% | 7568 | 1.3602 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
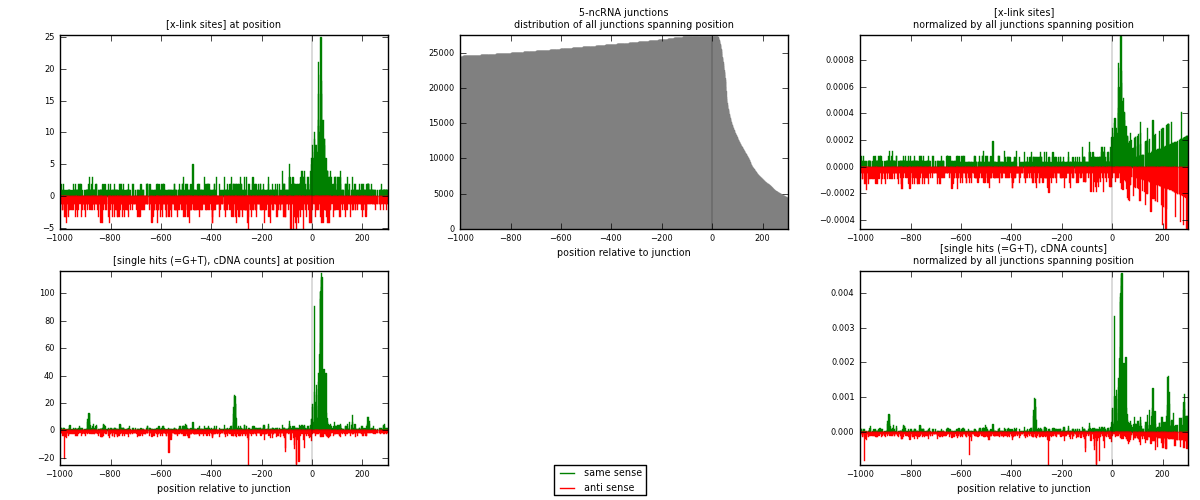 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1355 | 0.98% | 3.05% | 52.18% | 636 | 1.02964 |
| anti | 1242 | 0.90% | 2.80% | 47.82% | 693 | 0.943769 |
| both | 2597 | 1.88% | 5.84% | 100.00% | 1316 | 1.9734 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3128 | 2.01% | 6.04% | 67.20% | 636 | 2.3769 |
| anti | 1527 | 0.98% | 2.95% | 32.80% | 693 | 1.16033 |
| both | 4655 | 3.00% | 8.98% | 100.00% | 1316 | 3.53723 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
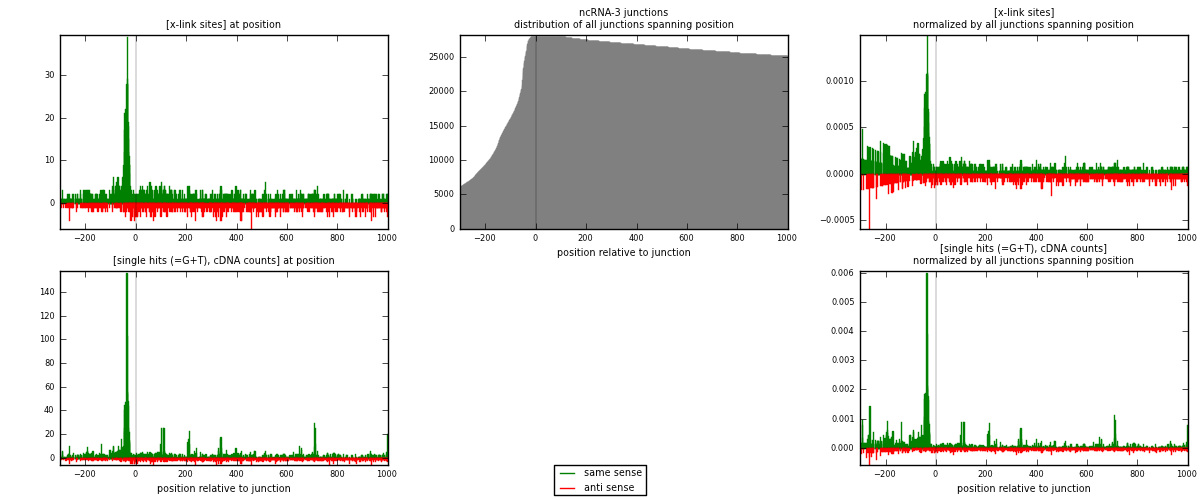 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1724 | 1.25% | 3.88% | 63.52% | 785 | 1.24207 |
| anti | 990 | 0.72% | 2.23% | 36.48% | 615 | 0.713256 |
| both | 2714 | 1.97% | 6.11% | 100.00% | 1388 | 1.95533 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2944 | 1.89% | 5.68% | 73.38% | 785 | 2.12104 |
| anti | 1068 | 0.69% | 2.06% | 26.62% | 615 | 0.769452 |
| both | 4012 | 2.58% | 7.74% | 100.00% | 1388 | 2.89049 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
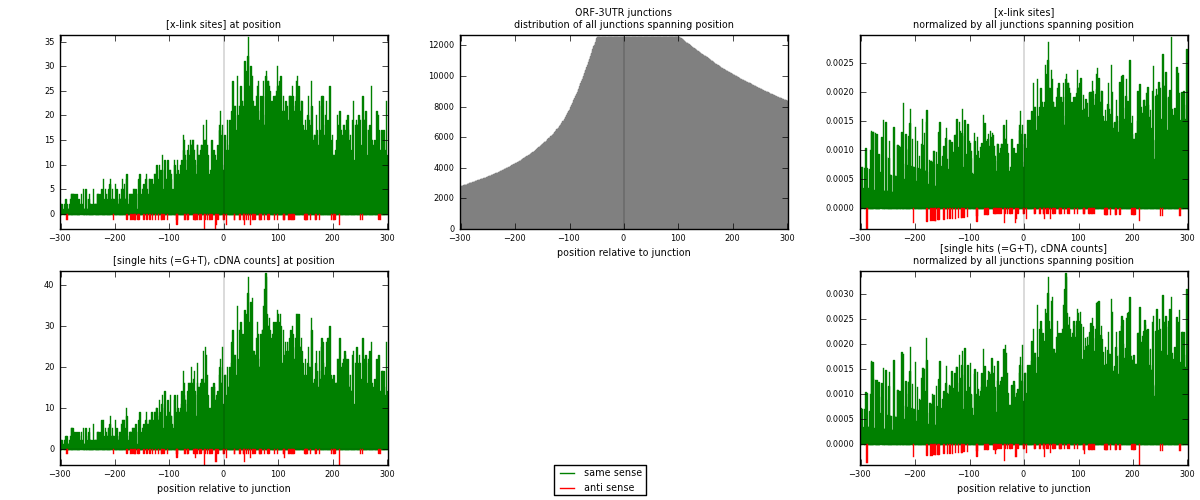
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7204 | 5.22% | 16.21% | 98.55% | 2596 | 2.71439 |
| anti | 106 | 0.08% | 0.24% | 1.45% | 71 | 0.0399397 |
| both | 7310 | 5.29% | 16.45% | 100.00% | 2654 | 2.75433 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 8339 | 5.37% | 16.09% | 98.66% | 2596 | 3.14205 |
| anti | 113 | 0.07% | 0.22% | 1.34% | 71 | 0.0425772 |
| both | 8452 | 5.44% | 16.31% | 100.00% | 2654 | 3.18463 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
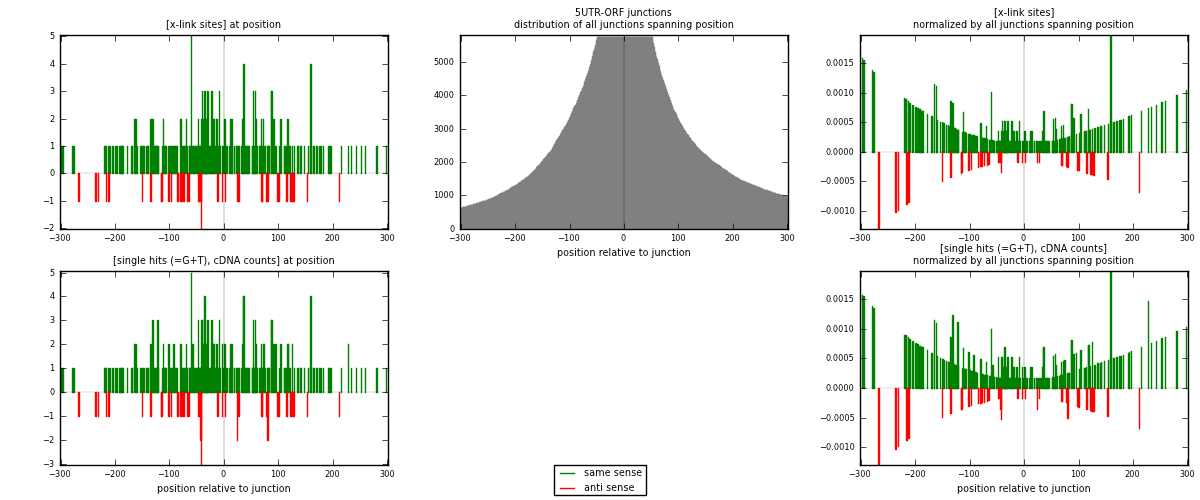
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 245 | 0.18% | 0.55% | 85.66% | 184 | 1.2069 |
| anti | 41 | 0.03% | 0.09% | 14.34% | 21 | 0.20197 |
| both | 286 | 0.21% | 0.64% | 100.00% | 203 | 1.40887 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 263 | 0.17% | 0.51% | 85.39% | 184 | 1.29557 |
| anti | 45 | 0.03% | 0.09% | 14.61% | 21 | 0.221675 |
| both | 308 | 0.20% | 0.59% | 100.00% | 203 | 1.51724 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
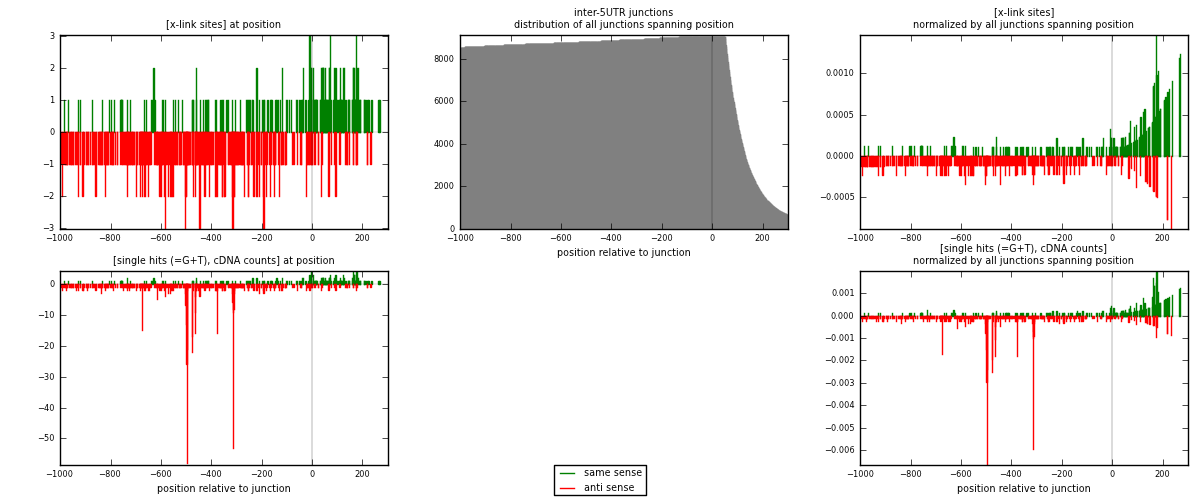
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 212 | 0.15% | 0.48% | 33.92% | 156 | 0.486239 |
| anti | 413 | 0.30% | 0.93% | 66.08% | 291 | 0.947248 |
| both | 625 | 0.45% | 1.41% | 100.00% | 436 | 1.43349 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 233 | 0.15% | 0.45% | 24.10% | 156 | 0.534404 |
| anti | 734 | 0.47% | 1.42% | 75.90% | 291 | 1.68349 |
| both | 967 | 0.62% | 1.87% | 100.00% | 436 | 2.21789 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
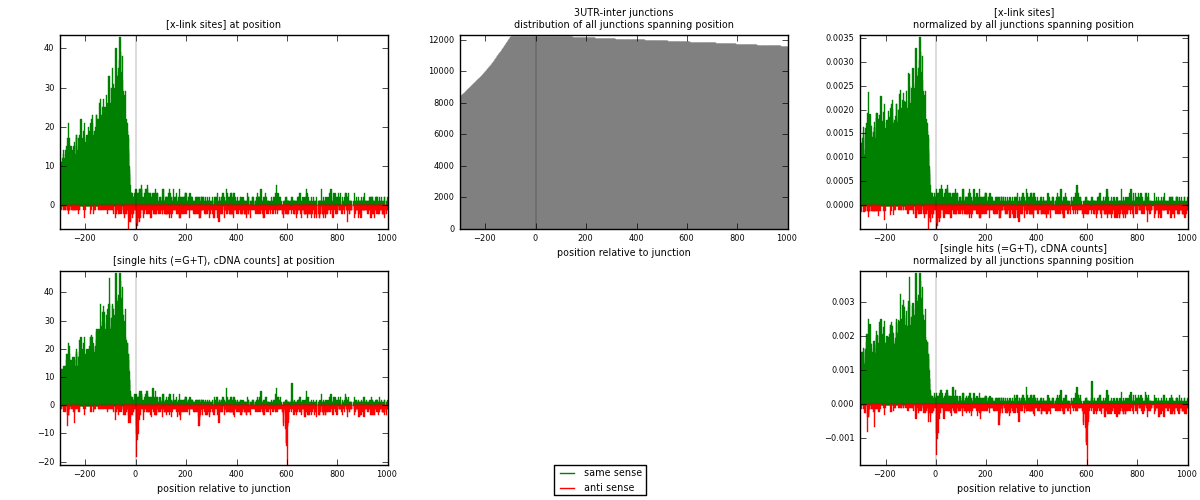
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5867 | 4.25% | 13.20% | 85.24% | 2457 | 2.10891 |
| anti | 1016 | 0.74% | 2.29% | 14.76% | 411 | 0.365205 |
| both | 6883 | 4.98% | 15.49% | 100.00% | 2782 | 2.47412 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 6561 | 4.22% | 12.66% | 83.45% | 2457 | 2.35838 |
| anti | 1301 | 0.84% | 2.51% | 16.55% | 411 | 0.467649 |
| both | 7862 | 5.06% | 15.17% | 100.00% | 2782 | 2.82602 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
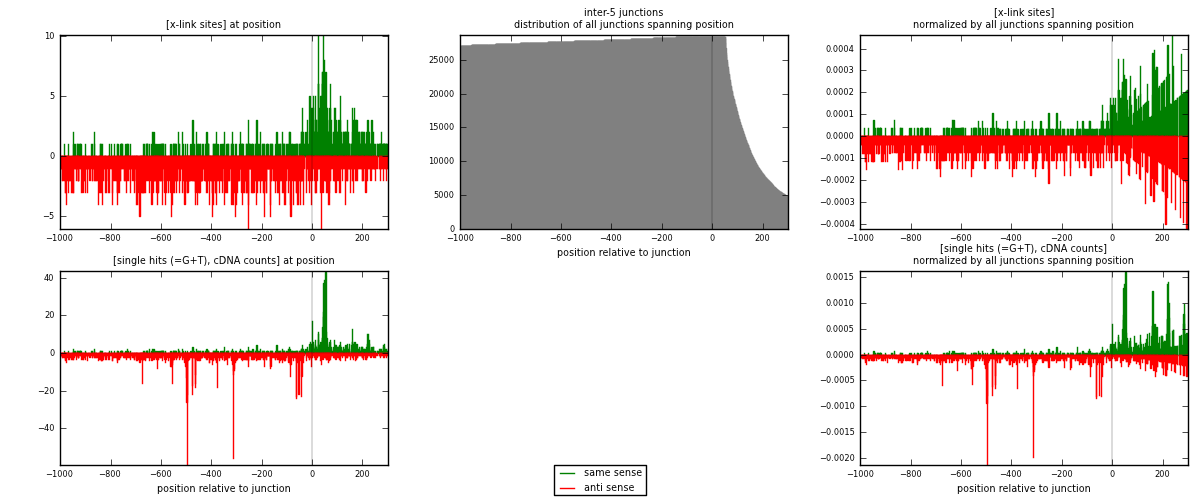 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 744 | 0.54% | 1.67% | 32.33% | 429 | 0.575851 |
| anti | 1557 | 1.13% | 3.50% | 67.67% | 884 | 1.20511 |
| both | 2301 | 1.67% | 5.18% | 100.00% | 1292 | 1.78096 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1141 | 0.73% | 2.20% | 34.67% | 429 | 0.883127 |
| anti | 2150 | 1.38% | 4.15% | 65.33% | 884 | 1.66409 |
| both | 3291 | 2.12% | 6.35% | 100.00% | 1292 | 2.54721 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
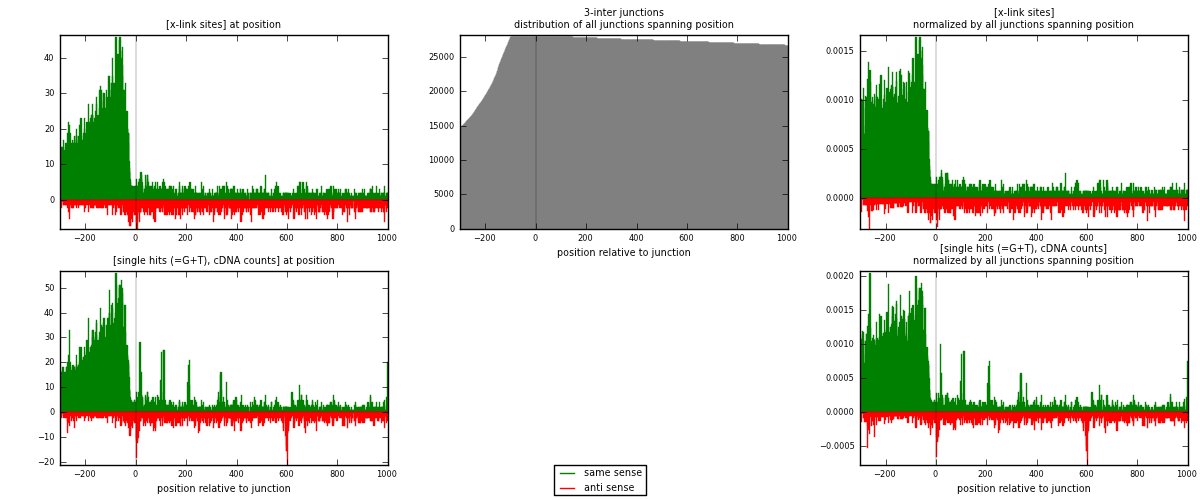 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7319 | 5.30% | 16.47% | 79.81% | 3008 | 1.96062 |
| anti | 1851 | 1.34% | 4.17% | 20.19% | 831 | 0.495848 |
| both | 9170 | 6.64% | 20.64% | 100.00% | 3733 | 2.45647 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 8853 | 5.70% | 17.08% | 79.84% | 3008 | 2.37155 |
| anti | 2235 | 1.44% | 4.31% | 20.16% | 831 | 0.598714 |
| both | 11088 | 7.14% | 21.40% | 100.00% | 3733 | 2.97027 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
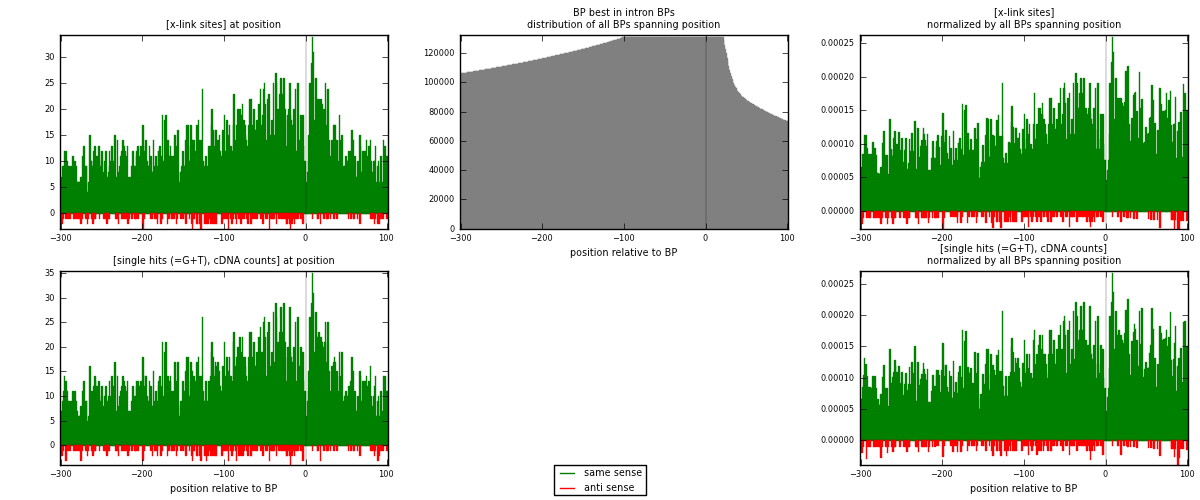 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5063 | 3.67% | 11.39% | 95.65% | 4425 | 1.09707 |
| anti | 230 | 0.17% | 0.52% | 4.35% | 197 | 0.0498375 |
| both | 5293 | 3.83% | 11.91% | 100.00% | 4615 | 1.14691 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 5290 | 3.40% | 10.21% | 95.63% | 4425 | 1.14626 |
| anti | 242 | 0.16% | 0.47% | 4.37% | 197 | 0.0524377 |
| both | 5532 | 3.56% | 10.68% | 100.00% | 4615 | 1.1987 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.