RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
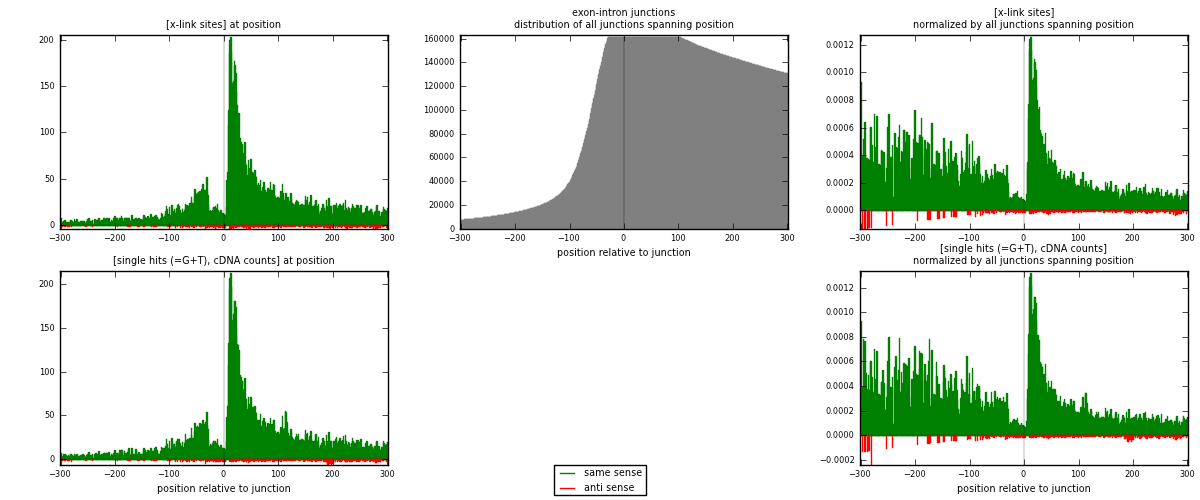 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13723 | 9.34% | 28.39% | 97.70% | 10731 | 1.25416 |
| anti | 323 | 0.22% | 0.67% | 2.30% | 229 | 0.0295193 |
| both | 14046 | 9.56% | 29.06% | 100.00% | 10942 | 1.28368 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 14391 | 8.95% | 26.54% | 97.39% | 10731 | 1.31521 |
| anti | 385 | 0.24% | 0.71% | 2.61% | 229 | 0.0351855 |
| both | 14776 | 9.19% | 27.25% | 100.00% | 10942 | 1.35039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
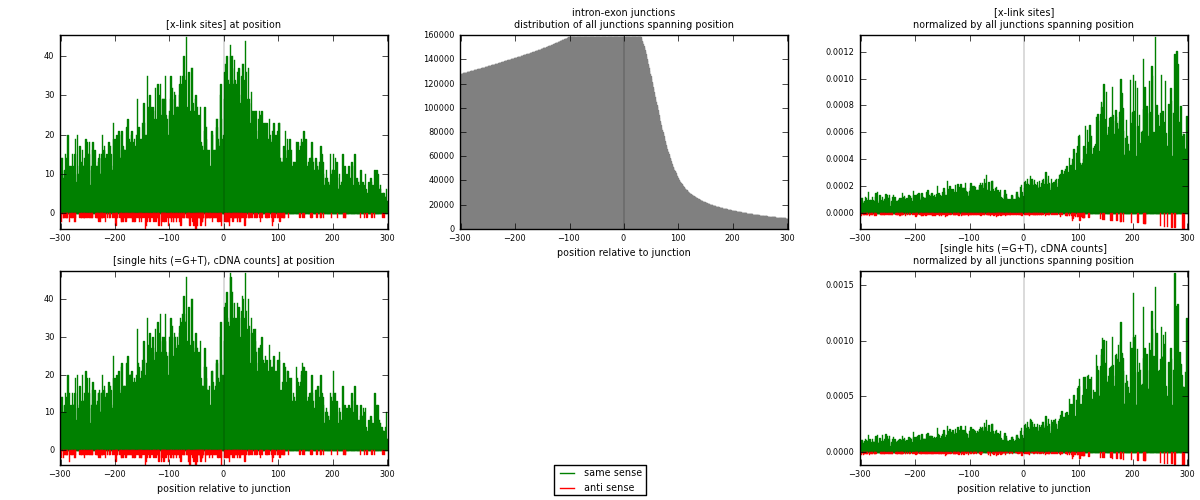 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10065 | 6.85% | 20.82% | 96.73% | 7743 | 1.25655 |
| anti | 340 | 0.23% | 0.70% | 3.27% | 289 | 0.0424469 |
| both | 10405 | 7.08% | 21.52% | 100.00% | 8010 | 1.299 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 10693 | 6.65% | 19.72% | 96.80% | 7743 | 1.33496 |
| anti | 353 | 0.22% | 0.65% | 3.20% | 289 | 0.0440699 |
| both | 11046 | 6.87% | 20.37% | 100.00% | 8010 | 1.37903 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
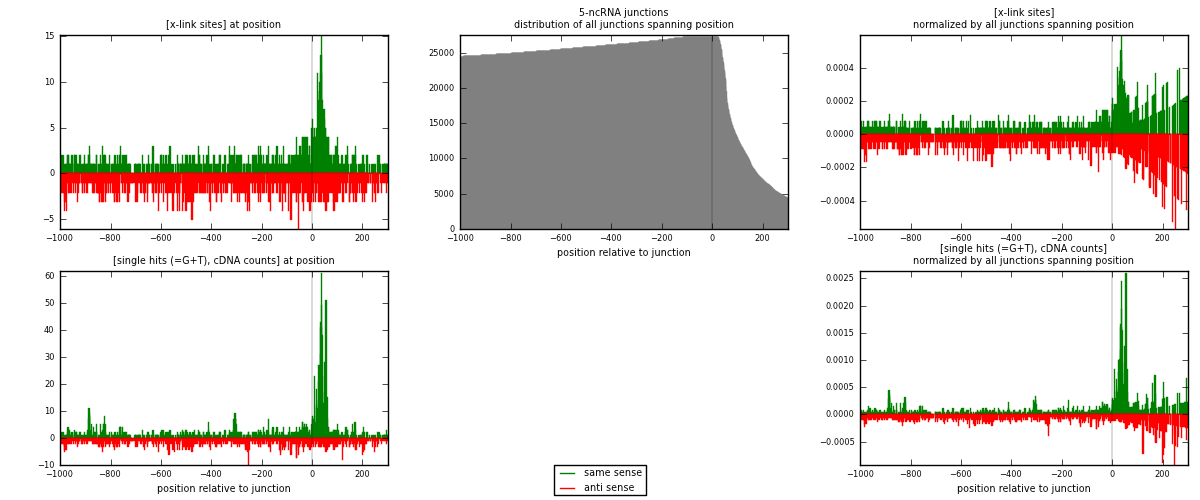 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1110 | 0.76% | 2.30% | 46.46% | 516 | 0.935919 |
| anti | 1279 | 0.87% | 2.65% | 53.54% | 678 | 1.07841 |
| both | 2389 | 1.63% | 4.94% | 100.00% | 1186 | 2.01433 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1915 | 1.19% | 3.53% | 57.35% | 516 | 1.61467 |
| anti | 1424 | 0.89% | 2.63% | 42.65% | 678 | 1.20067 |
| both | 3339 | 2.08% | 6.16% | 100.00% | 1186 | 2.81535 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
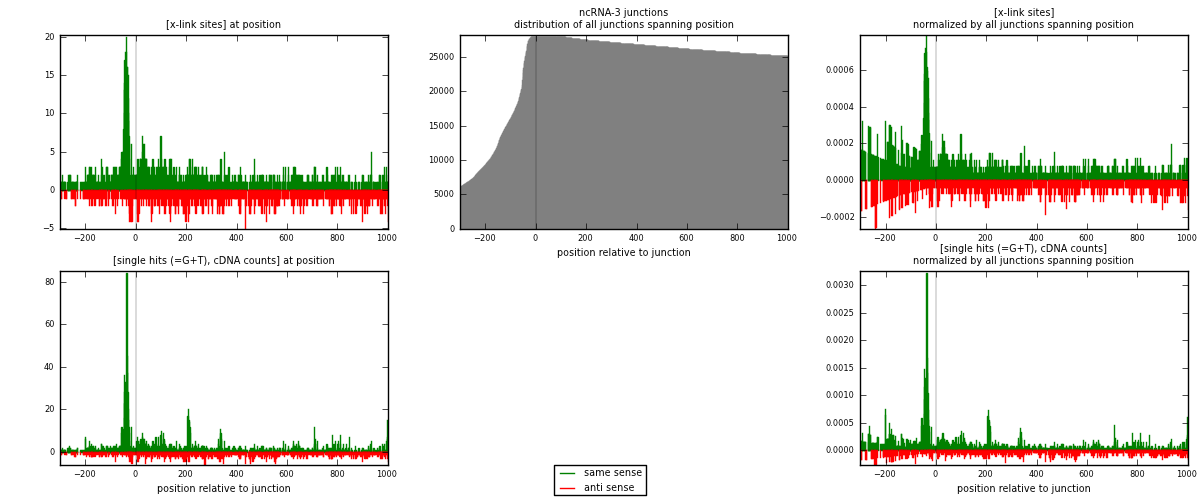 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1590 | 1.08% | 3.29% | 61.32% | 755 | 1.21839 |
| anti | 1003 | 0.68% | 2.07% | 38.68% | 563 | 0.768582 |
| both | 2593 | 1.76% | 5.36% | 100.00% | 1305 | 1.98697 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2307 | 1.44% | 4.25% | 67.87% | 755 | 1.76782 |
| anti | 1092 | 0.68% | 2.01% | 32.13% | 563 | 0.836782 |
| both | 3399 | 2.12% | 6.27% | 100.00% | 1305 | 2.6046 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
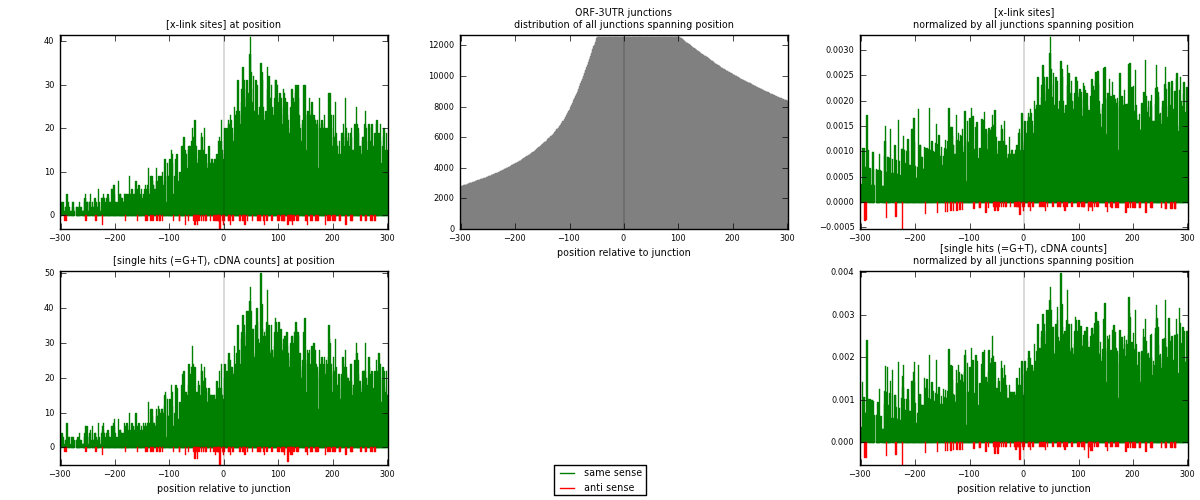
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8167 | 5.56% | 16.89% | 98.50% | 2659 | 2.99706 |
| anti | 124 | 0.08% | 0.26% | 1.50% | 76 | 0.0455046 |
| both | 8291 | 5.64% | 17.15% | 100.00% | 2725 | 3.04257 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 9538 | 5.94% | 17.59% | 98.62% | 2659 | 3.50018 |
| anti | 133 | 0.08% | 0.25% | 1.38% | 76 | 0.0488073 |
| both | 9671 | 6.02% | 17.83% | 100.00% | 2725 | 3.54899 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
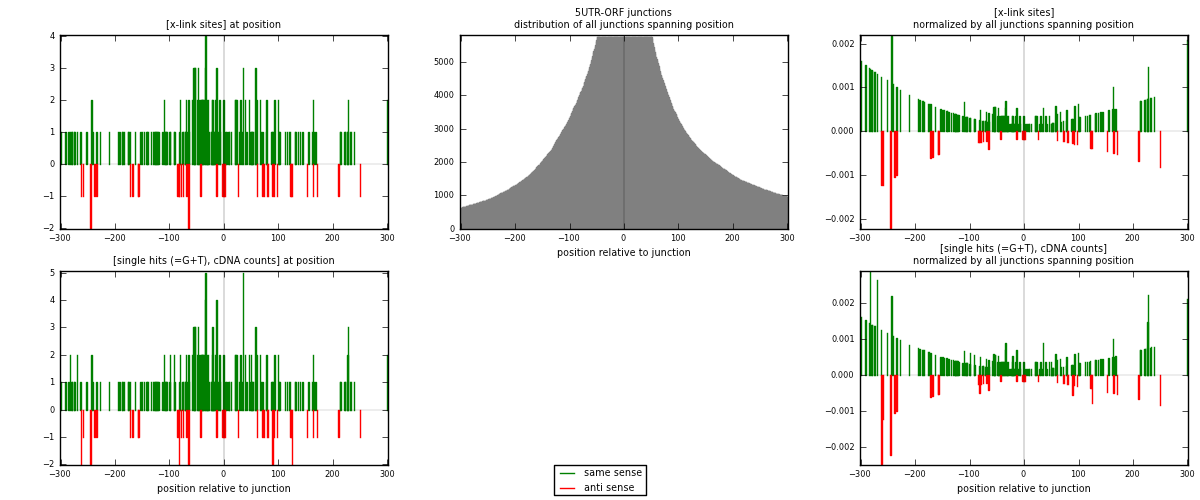
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 211 | 0.14% | 0.44% | 83.73% | 146 | 1.30247 |
| anti | 41 | 0.03% | 0.08% | 16.27% | 18 | 0.253086 |
| both | 252 | 0.17% | 0.52% | 100.00% | 162 | 1.55556 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 224 | 0.14% | 0.41% | 83.27% | 146 | 1.38272 |
| anti | 45 | 0.03% | 0.08% | 16.73% | 18 | 0.277778 |
| both | 269 | 0.17% | 0.50% | 100.00% | 162 | 1.66049 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
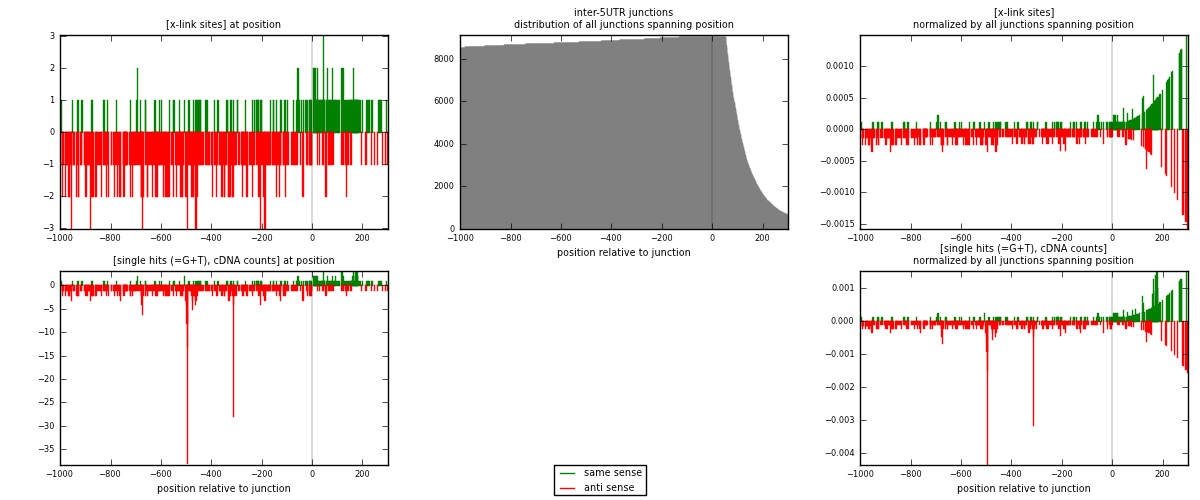
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 177 | 0.12% | 0.37% | 27.15% | 131 | 0.409722 |
| anti | 475 | 0.32% | 0.98% | 72.85% | 311 | 1.09954 |
| both | 652 | 0.44% | 1.35% | 100.00% | 432 | 1.50926 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 192 | 0.12% | 0.35% | 24.27% | 131 | 0.444444 |
| anti | 599 | 0.37% | 1.10% | 75.73% | 311 | 1.38657 |
| both | 791 | 0.49% | 1.46% | 100.00% | 432 | 1.83102 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
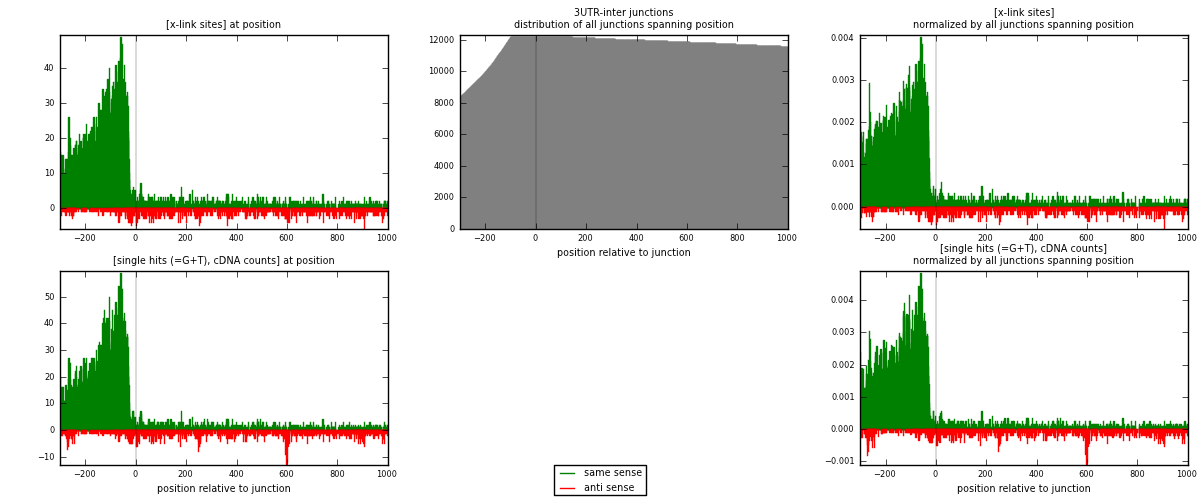
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6791 | 4.62% | 14.05% | 85.56% | 2574 | 2.31854 |
| anti | 1146 | 0.78% | 2.37% | 14.44% | 434 | 0.39126 |
| both | 7937 | 5.40% | 16.42% | 100.00% | 2929 | 2.7098 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 7646 | 4.76% | 14.10% | 84.90% | 2574 | 2.61045 |
| anti | 1360 | 0.85% | 2.51% | 15.10% | 434 | 0.464322 |
| both | 9006 | 5.60% | 16.61% | 100.00% | 2929 | 3.07477 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
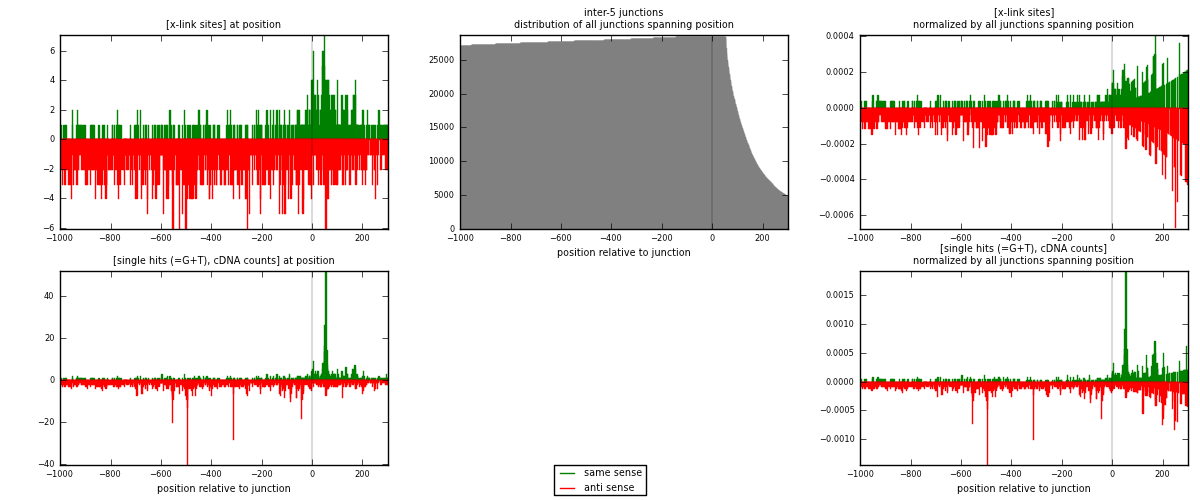
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 551 | 0.37% | 1.14% | 25.44% | 324 | 0.464587 |
| anti | 1615 | 1.10% | 3.34% | 74.56% | 879 | 1.36172 |
| both | 2166 | 1.47% | 4.48% | 100.00% | 1186 | 1.82631 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 766 | 0.48% | 1.41% | 28.10% | 324 | 0.645868 |
| anti | 1960 | 1.22% | 3.61% | 71.90% | 879 | 1.65261 |
| both | 2726 | 1.70% | 5.03% | 100.00% | 1186 | 2.29848 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
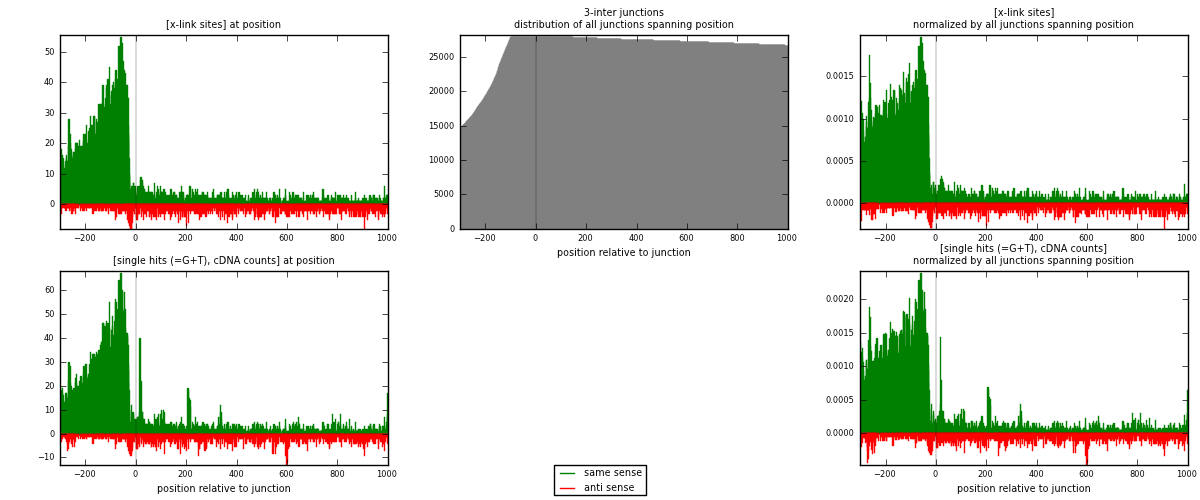
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8304 | 5.65% | 17.18% | 80.74% | 3133 | 2.15018 |
| anti | 1981 | 1.35% | 4.10% | 19.26% | 826 | 0.512947 |
| both | 10285 | 7.00% | 21.28% | 100.00% | 3862 | 2.66313 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 9843 | 6.12% | 18.15% | 80.98% | 3133 | 2.54868 |
| anti | 2312 | 1.44% | 4.26% | 19.02% | 826 | 0.598654 |
| both | 12155 | 7.56% | 22.41% | 100.00% | 3862 | 3.14733 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
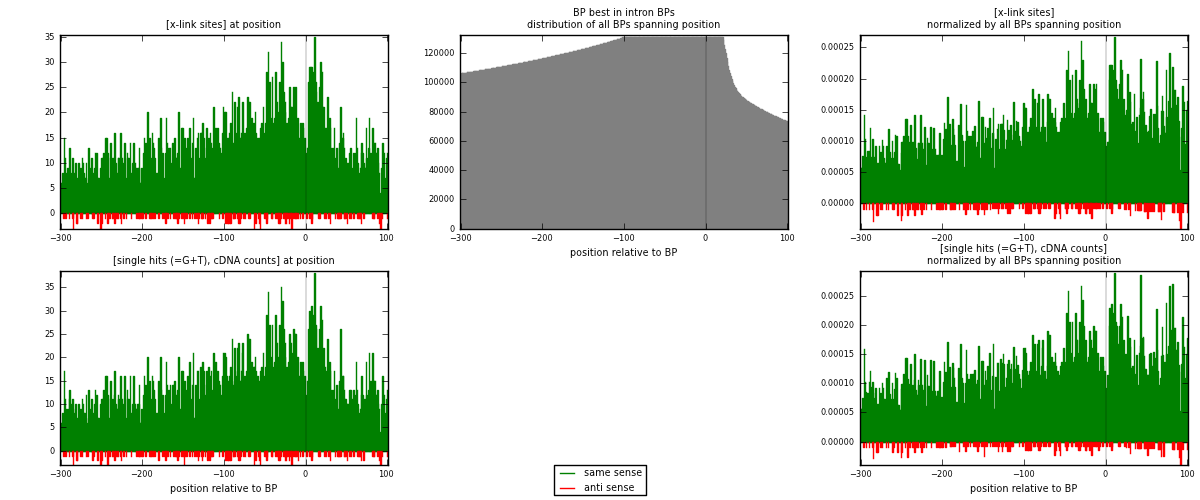
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5386 | 3.66% | 11.14% | 96.21% | 4596 | 1.13056 |
| anti | 212 | 0.14% | 0.44% | 3.79% | 173 | 0.0445004 |
| both | 5598 | 3.81% | 11.58% | 100.00% | 4764 | 1.17506 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 5606 | 3.49% | 10.34% | 96.24% | 4596 | 1.17674 |
| anti | 219 | 0.14% | 0.40% | 3.76% | 173 | 0.0459698 |
| both | 5825 | 3.62% | 10.74% | 100.00% | 4764 | 1.22271 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.