RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
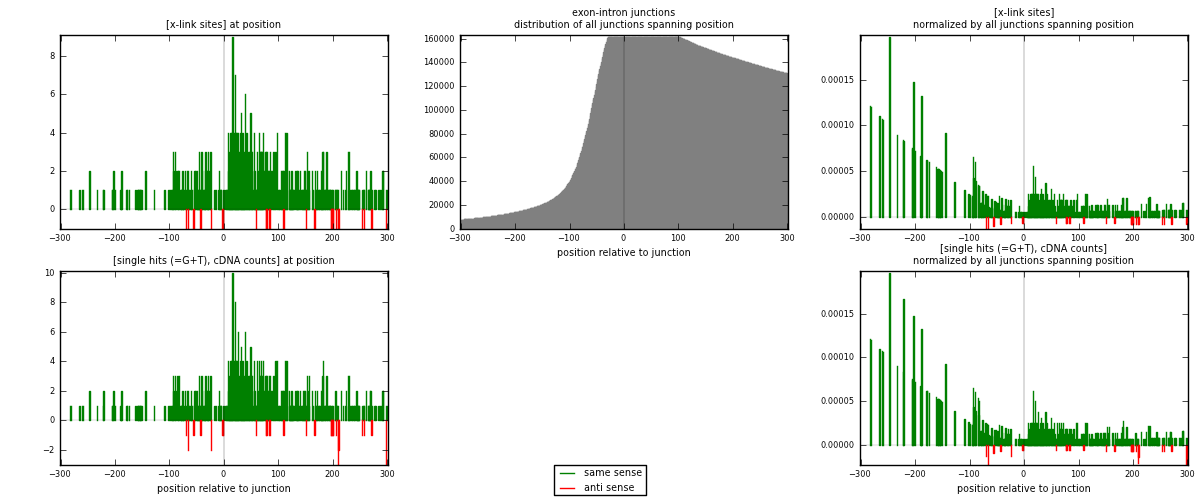 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 509 | 7.08% | 23.51% | 94.96% | 479 | 1.02209 |
| anti | 27 | 0.38% | 1.25% | 5.04% | 19 | 0.0542169 |
| both | 536 | 7.46% | 24.76% | 100.00% | 498 | 1.07631 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 525 | 6.94% | 23.37% | 93.92% | 479 | 1.05422 |
| anti | 34 | 0.45% | 1.51% | 6.08% | 19 | 0.0682731 |
| both | 559 | 7.39% | 24.89% | 100.00% | 498 | 1.12249 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
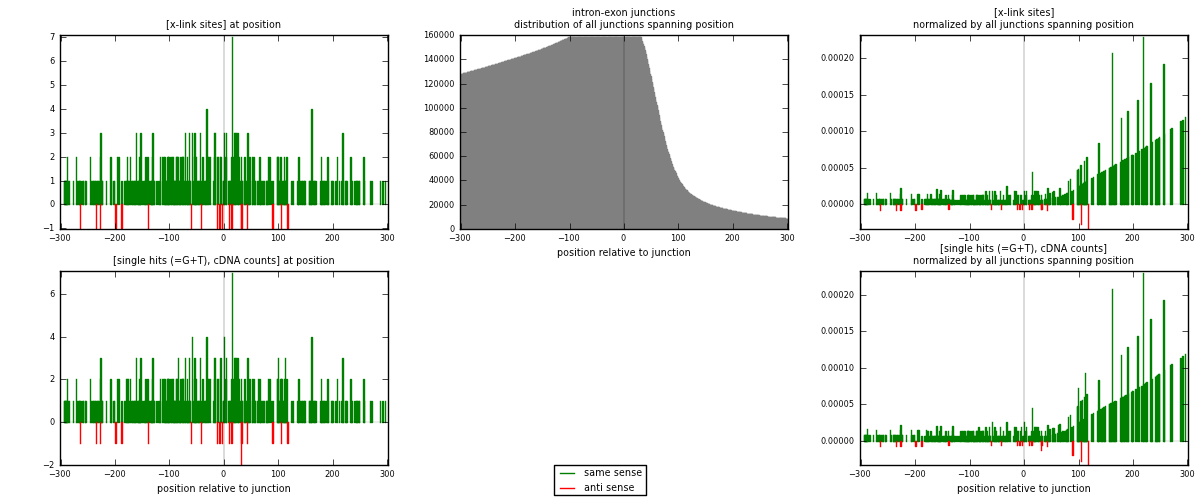 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 404 | 5.62% | 18.66% | 95.51% | 367 | 1.05208 |
| anti | 19 | 0.26% | 0.88% | 4.49% | 17 | 0.0494792 |
| both | 423 | 5.88% | 19.54% | 100.00% | 384 | 1.10156 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 417 | 5.51% | 18.57% | 95.42% | 367 | 1.08594 |
| anti | 20 | 0.26% | 0.89% | 4.58% | 17 | 0.0520833 |
| both | 437 | 5.77% | 19.46% | 100.00% | 384 | 1.13802 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
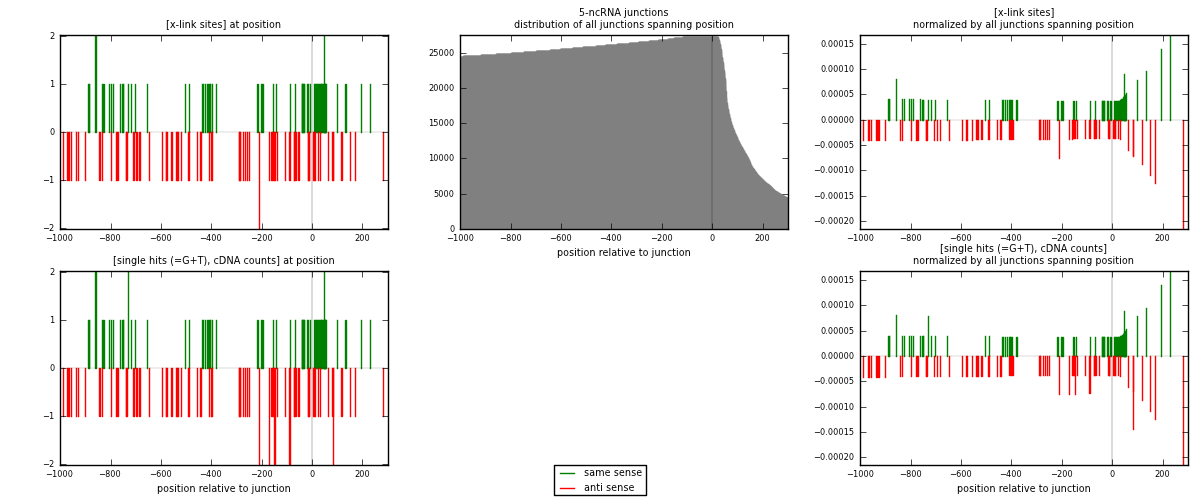 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 64 | 0.89% | 2.96% | 47.76% | 34 | 0.666667 |
| anti | 70 | 0.97% | 3.23% | 52.24% | 62 | 0.729167 |
| both | 134 | 1.86% | 6.19% | 100.00% | 96 | 1.39583 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 65 | 0.86% | 2.89% | 46.76% | 34 | 0.677083 |
| anti | 74 | 0.98% | 3.29% | 53.24% | 62 | 0.770833 |
| both | 139 | 1.84% | 6.19% | 100.00% | 96 | 1.44792 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
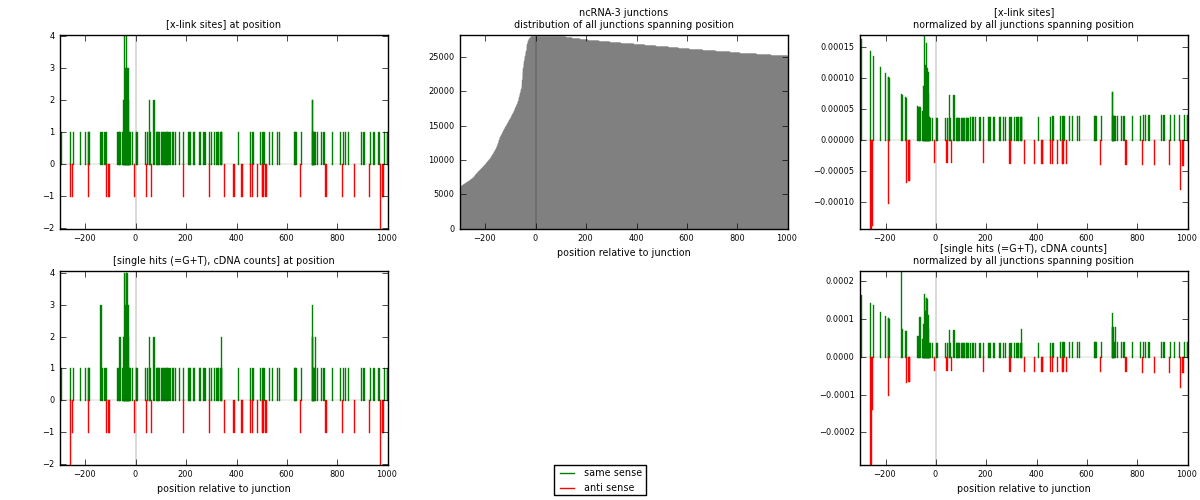 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 145 | 2.02% | 6.70% | 82.86% | 98 | 1.14173 |
| anti | 30 | 0.42% | 1.39% | 17.14% | 29 | 0.23622 |
| both | 175 | 2.43% | 8.08% | 100.00% | 127 | 1.37795 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 154 | 2.03% | 6.86% | 83.24% | 98 | 1.2126 |
| anti | 31 | 0.41% | 1.38% | 16.76% | 29 | 0.244094 |
| both | 185 | 2.44% | 8.24% | 100.00% | 127 | 1.45669 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
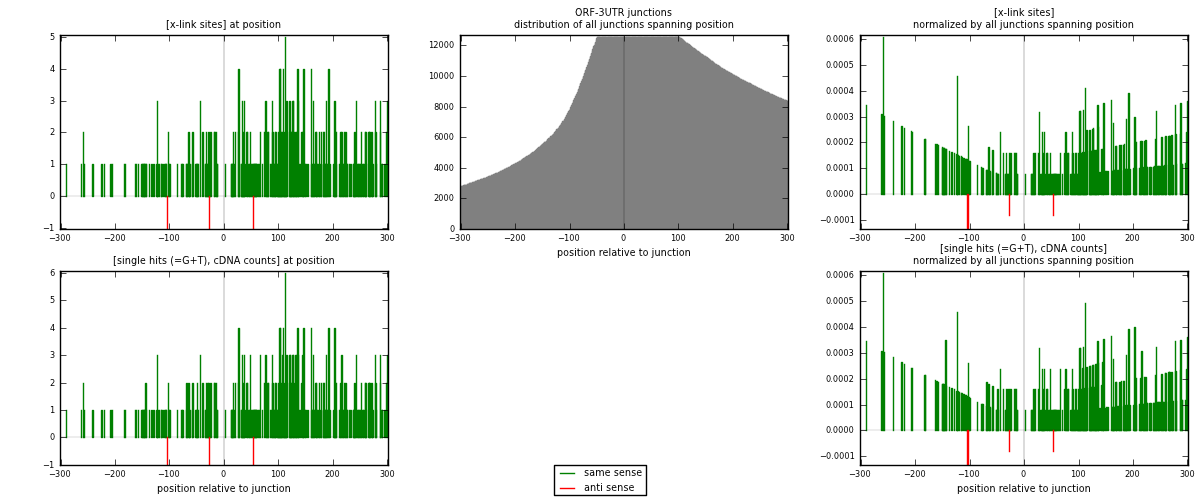
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 374 | 5.20% | 17.27% | 99.20% | 288 | 1.28522 |
| anti | 3 | 0.04% | 0.14% | 0.80% | 3 | 0.0103093 |
| both | 377 | 5.24% | 17.41% | 100.00% | 291 | 1.29553 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 388 | 5.13% | 17.28% | 99.23% | 288 | 1.33333 |
| anti | 3 | 0.04% | 0.13% | 0.77% | 3 | 0.0103093 |
| both | 391 | 5.17% | 17.41% | 100.00% | 291 | 1.34364 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
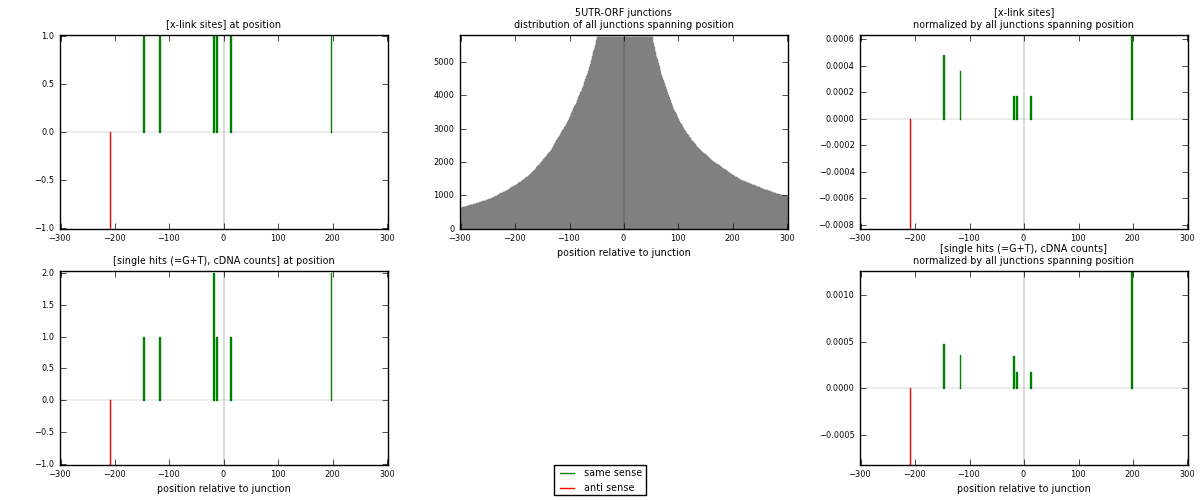
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6 | 0.08% | 0.28% | 85.71% | 6 | 0.857143 |
| anti | 1 | 0.01% | 0.05% | 14.29% | 1 | 0.142857 |
| both | 7 | 0.10% | 0.32% | 100.00% | 7 | 1 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 8 | 0.11% | 0.36% | 88.89% | 6 | 1.14286 |
| anti | 1 | 0.01% | 0.04% | 11.11% | 1 | 0.142857 |
| both | 9 | 0.12% | 0.40% | 100.00% | 7 | 1.28571 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
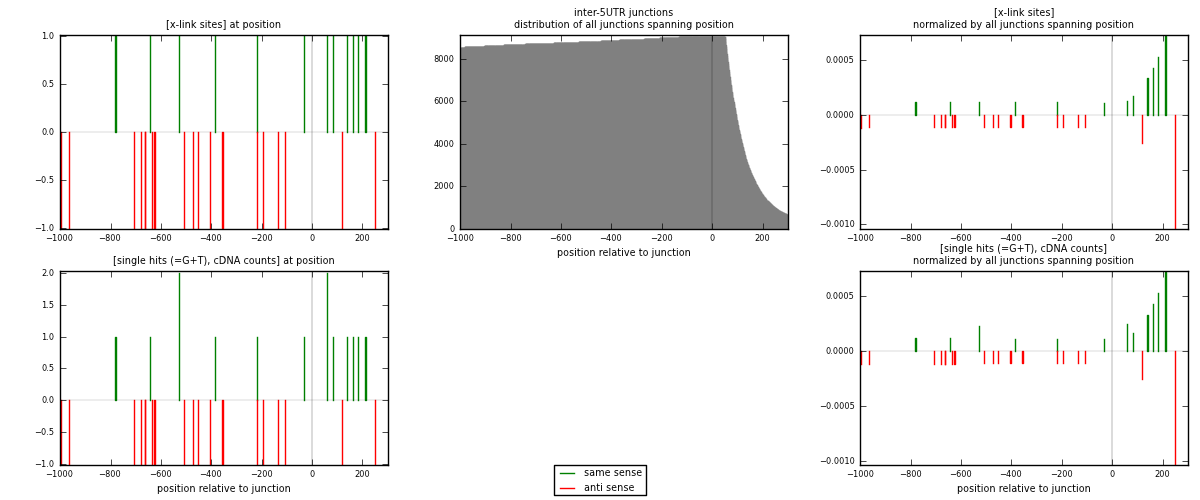
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12 | 0.17% | 0.55% | 38.71% | 11 | 0.4 |
| anti | 19 | 0.26% | 0.88% | 61.29% | 19 | 0.633333 |
| both | 31 | 0.43% | 1.43% | 100.00% | 30 | 1.03333 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 14 | 0.18% | 0.62% | 42.42% | 11 | 0.466667 |
| anti | 19 | 0.25% | 0.85% | 57.58% | 19 | 0.633333 |
| both | 33 | 0.44% | 1.47% | 100.00% | 30 | 1.1 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
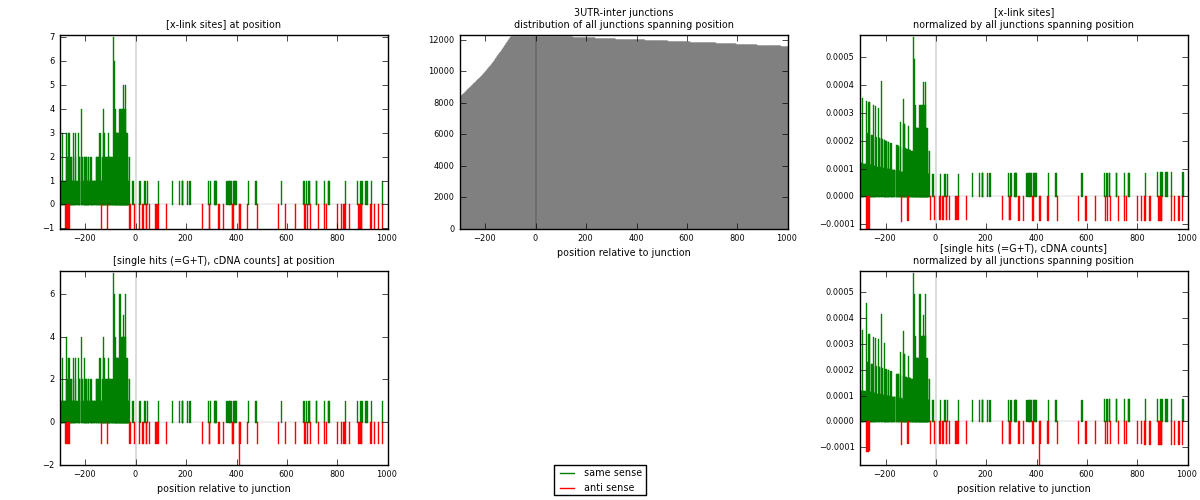
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 369 | 5.13% | 17.04% | 87.86% | 297 | 1.10149 |
| anti | 51 | 0.71% | 2.36% | 12.14% | 38 | 0.152239 |
| both | 420 | 5.84% | 19.40% | 100.00% | 335 | 1.25373 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 377 | 4.98% | 16.79% | 87.88% | 297 | 1.12537 |
| anti | 52 | 0.69% | 2.32% | 12.12% | 38 | 0.155224 |
| both | 429 | 5.67% | 19.10% | 100.00% | 335 | 1.2806 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
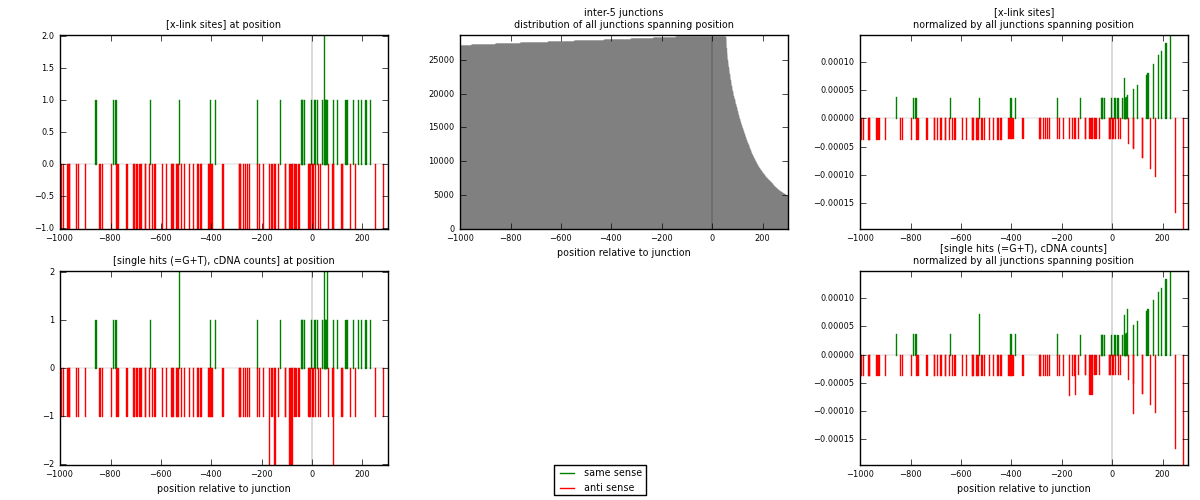
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 34 | 0.47% | 1.57% | 28.33% | 28 | 0.330097 |
| anti | 86 | 1.20% | 3.97% | 71.67% | 75 | 0.834951 |
| both | 120 | 1.67% | 5.54% | 100.00% | 103 | 1.16505 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 36 | 0.48% | 1.60% | 28.35% | 28 | 0.349515 |
| anti | 91 | 1.20% | 4.05% | 71.65% | 75 | 0.883495 |
| both | 127 | 1.68% | 5.65% | 100.00% | 103 | 1.23301 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
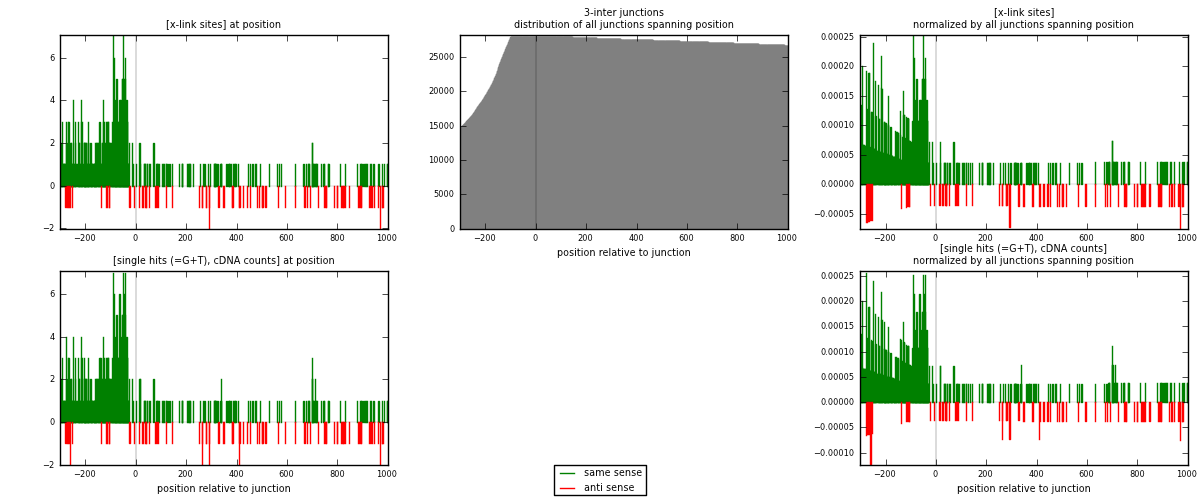
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 483 | 6.72% | 22.31% | 85.64% | 362 | 1.13647 |
| anti | 81 | 1.13% | 3.74% | 14.36% | 63 | 0.190588 |
| both | 564 | 7.85% | 26.05% | 100.00% | 425 | 1.32706 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 499 | 6.59% | 22.22% | 85.59% | 362 | 1.17412 |
| anti | 84 | 1.11% | 3.74% | 14.41% | 63 | 0.197647 |
| both | 583 | 7.70% | 25.96% | 100.00% | 425 | 1.37176 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
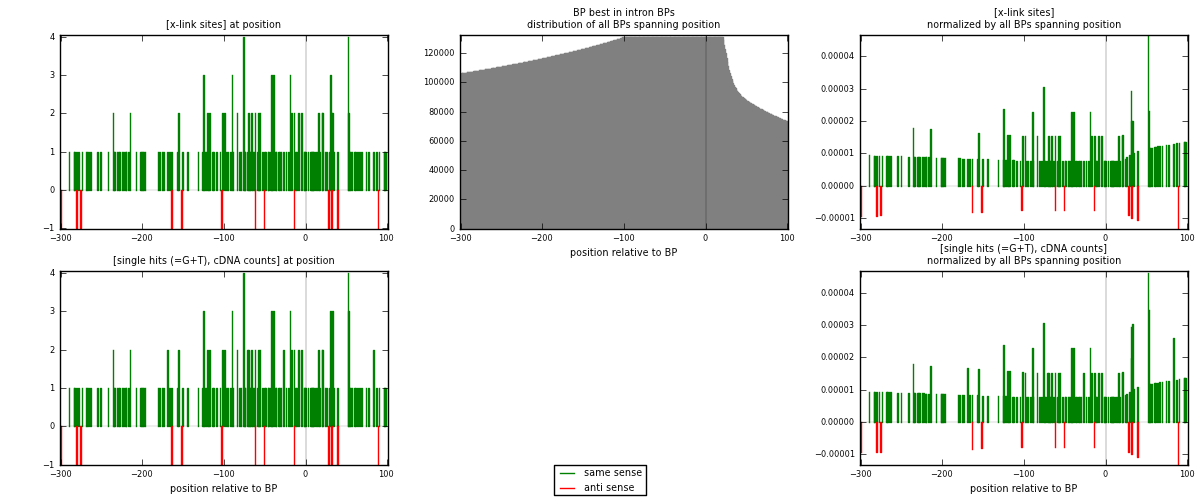
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 185 | 2.57% | 8.55% | 93.43% | 180 | 0.963542 |
| anti | 13 | 0.18% | 0.60% | 6.57% | 12 | 0.0677083 |
| both | 198 | 2.75% | 9.15% | 100.00% | 192 | 1.03125 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 191 | 2.52% | 8.50% | 93.63% | 180 | 0.994792 |
| anti | 13 | 0.17% | 0.58% | 6.37% | 12 | 0.0677083 |
| both | 204 | 2.70% | 9.08% | 100.00% | 192 | 1.0625 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.