RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
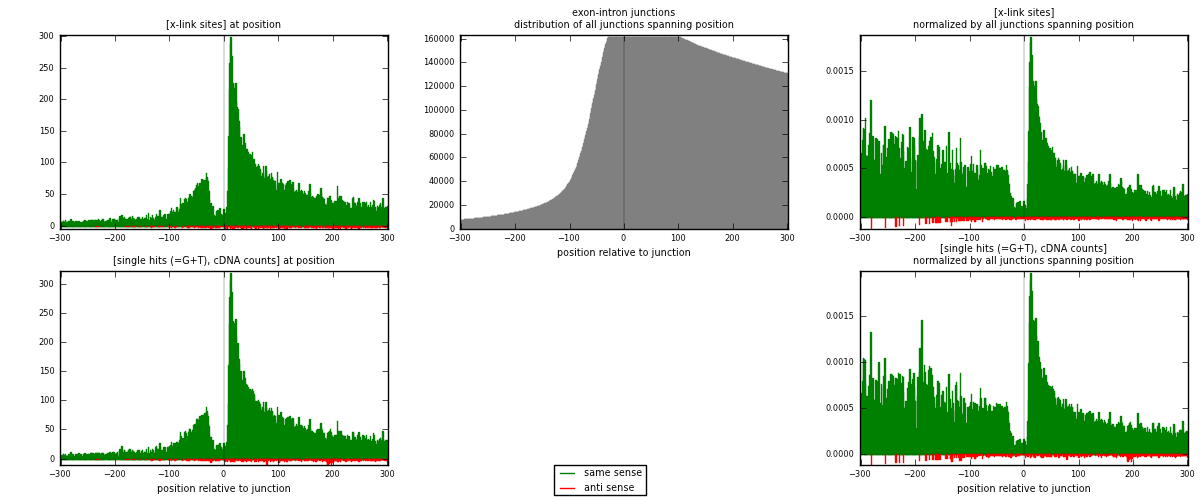 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 24819 | 9.12% | 30.47% | 97.50% | 17463 | 1.39011 |
| anti | 636 | 0.23% | 0.78% | 2.50% | 449 | 0.0356223 |
| both | 25455 | 9.35% | 31.25% | 100.00% | 17854 | 1.42573 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 26001 | 8.73% | 28.58% | 97.20% | 17463 | 1.45631 |
| anti | 750 | 0.25% | 0.82% | 2.80% | 449 | 0.0420074 |
| both | 26751 | 8.98% | 29.40% | 100.00% | 17854 | 1.49832 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
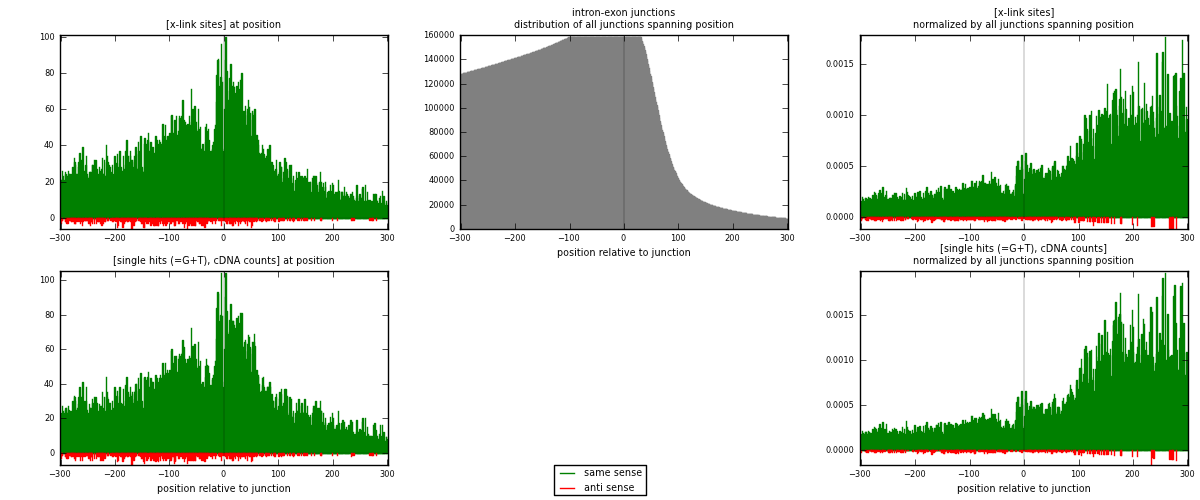 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 18610 | 6.84% | 22.85% | 96.67% | 13464 | 1.33491 |
| anti | 642 | 0.24% | 0.79% | 3.33% | 518 | 0.0460512 |
| both | 19252 | 7.07% | 23.63% | 100.00% | 13941 | 1.38096 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 19726 | 6.62% | 21.68% | 96.70% | 13464 | 1.41496 |
| anti | 674 | 0.23% | 0.74% | 3.30% | 518 | 0.0483466 |
| both | 20400 | 6.85% | 22.42% | 100.00% | 13941 | 1.46331 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
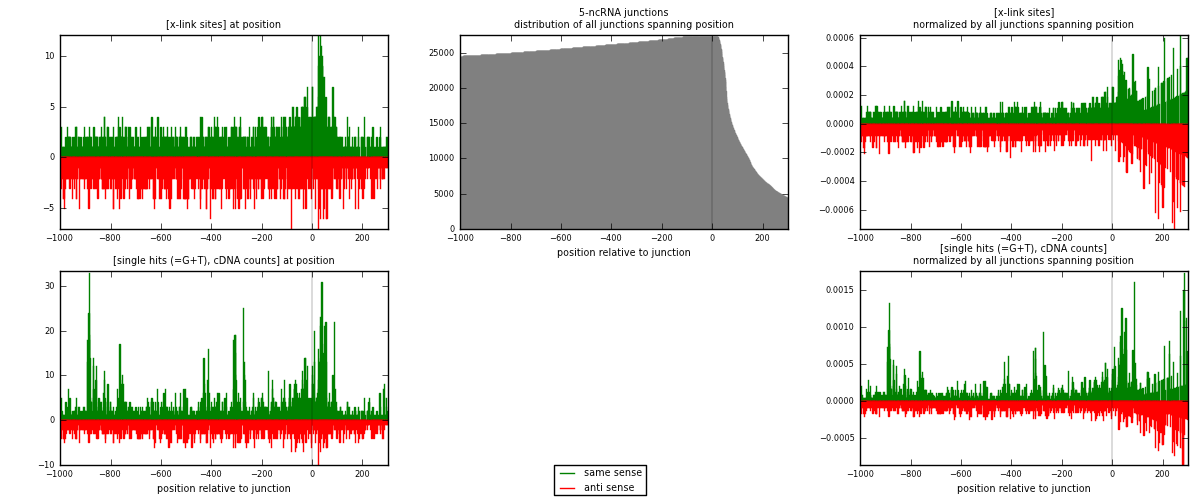 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1984 | 0.73% | 2.44% | 48.66% | 832 | 1.05983 |
| anti | 2093 | 0.77% | 2.57% | 51.34% | 1070 | 1.11806 |
| both | 4077 | 1.50% | 5.01% | 100.00% | 1872 | 2.17788 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3224 | 1.08% | 3.54% | 58.83% | 832 | 1.72222 |
| anti | 2256 | 0.76% | 2.48% | 41.17% | 1070 | 1.20513 |
| both | 5480 | 1.84% | 6.02% | 100.00% | 1872 | 2.92735 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
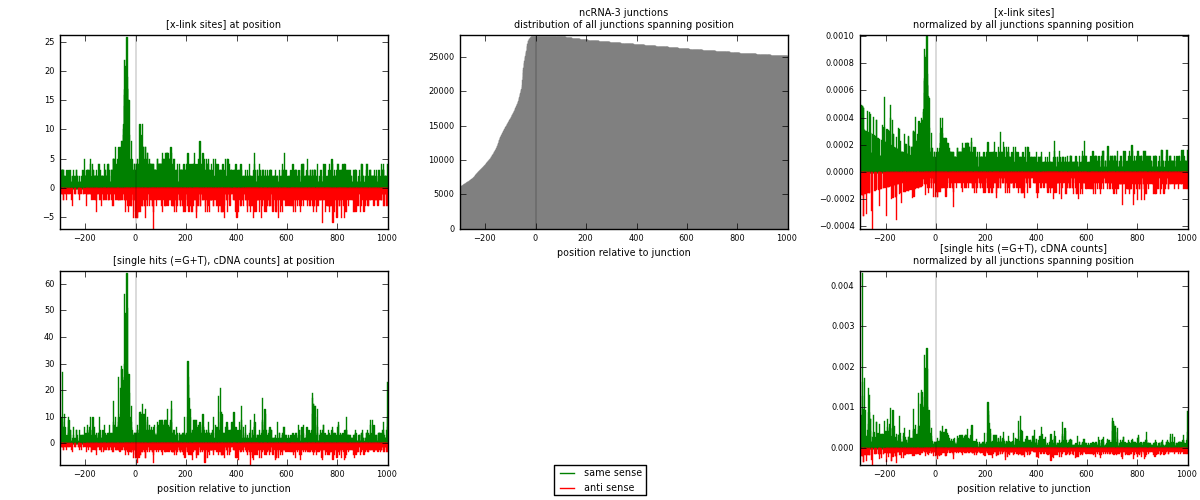 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2970 | 1.09% | 3.65% | 63.07% | 1231 | 1.39371 |
| anti | 1739 | 0.64% | 2.13% | 36.93% | 934 | 0.816049 |
| both | 4709 | 1.73% | 5.78% | 100.00% | 2131 | 2.20976 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4975 | 1.67% | 5.47% | 72.85% | 1231 | 2.33458 |
| anti | 1854 | 0.62% | 2.04% | 27.15% | 934 | 0.870014 |
| both | 6829 | 2.29% | 7.51% | 100.00% | 2131 | 3.2046 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
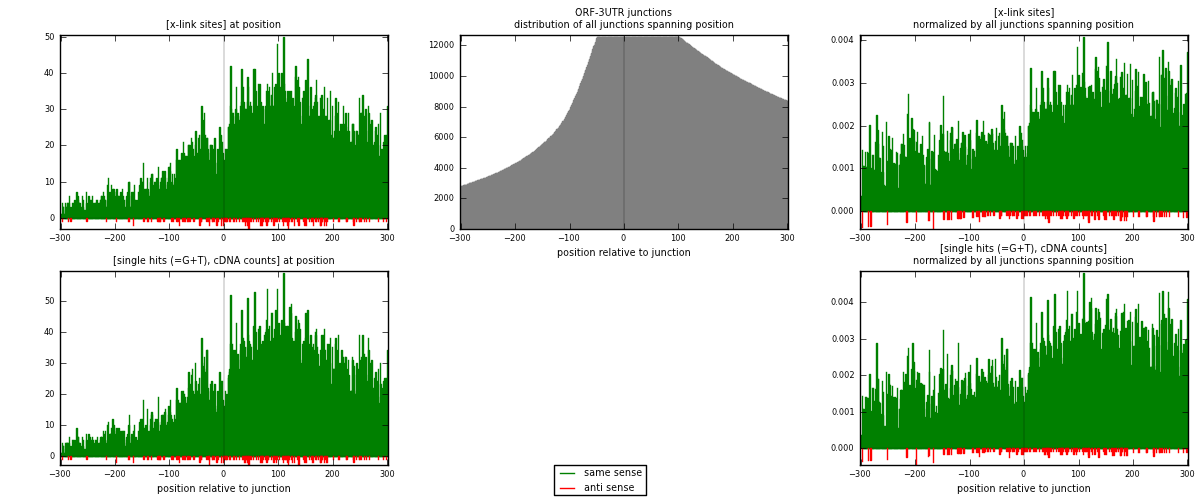
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 11003 | 4.04% | 13.51% | 98.50% | 3199 | 3.34539 |
| anti | 167 | 0.06% | 0.21% | 1.50% | 117 | 0.0507753 |
| both | 11170 | 4.10% | 13.71% | 100.00% | 3289 | 3.39617 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 12566 | 4.22% | 13.81% | 98.63% | 3199 | 3.82061 |
| anti | 175 | 0.06% | 0.19% | 1.37% | 117 | 0.0532077 |
| both | 12741 | 4.28% | 14.00% | 100.00% | 3289 | 3.87382 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
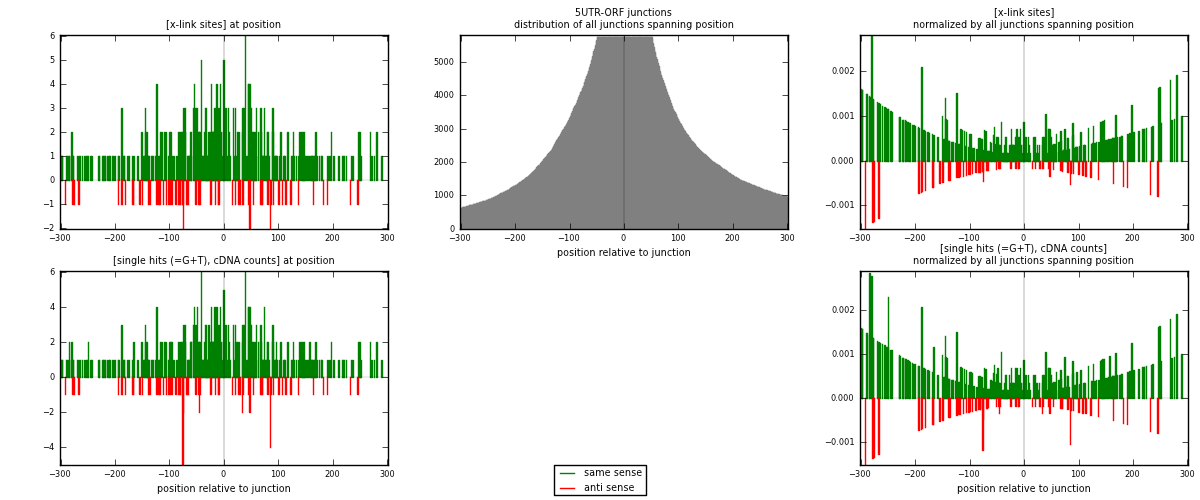
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 393 | 0.14% | 0.48% | 84.88% | 268 | 1.33673 |
| anti | 70 | 0.03% | 0.09% | 15.12% | 29 | 0.238095 |
| both | 463 | 0.17% | 0.57% | 100.00% | 294 | 1.57483 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 406 | 0.14% | 0.45% | 83.88% | 268 | 1.38095 |
| anti | 78 | 0.03% | 0.09% | 16.12% | 29 | 0.265306 |
| both | 484 | 0.16% | 0.53% | 100.00% | 294 | 1.64626 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
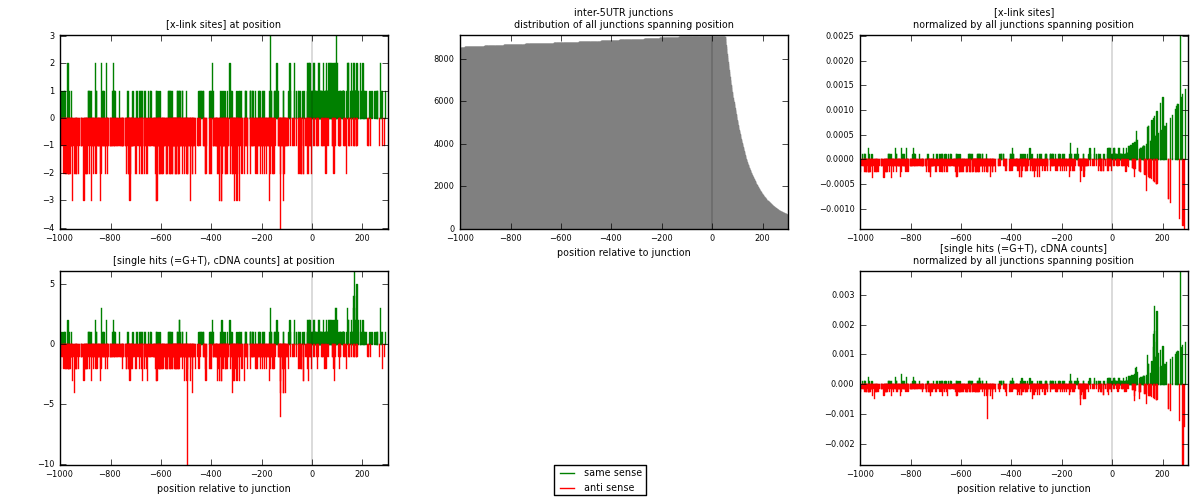
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 285 | 0.10% | 0.35% | 30.06% | 197 | 0.467213 |
| anti | 663 | 0.24% | 0.81% | 69.94% | 429 | 1.08689 |
| both | 948 | 0.35% | 1.16% | 100.00% | 610 | 1.5541 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 312 | 0.10% | 0.34% | 29.97% | 197 | 0.511475 |
| anti | 729 | 0.24% | 0.80% | 70.03% | 429 | 1.19508 |
| both | 1041 | 0.35% | 1.14% | 100.00% | 610 | 1.70656 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
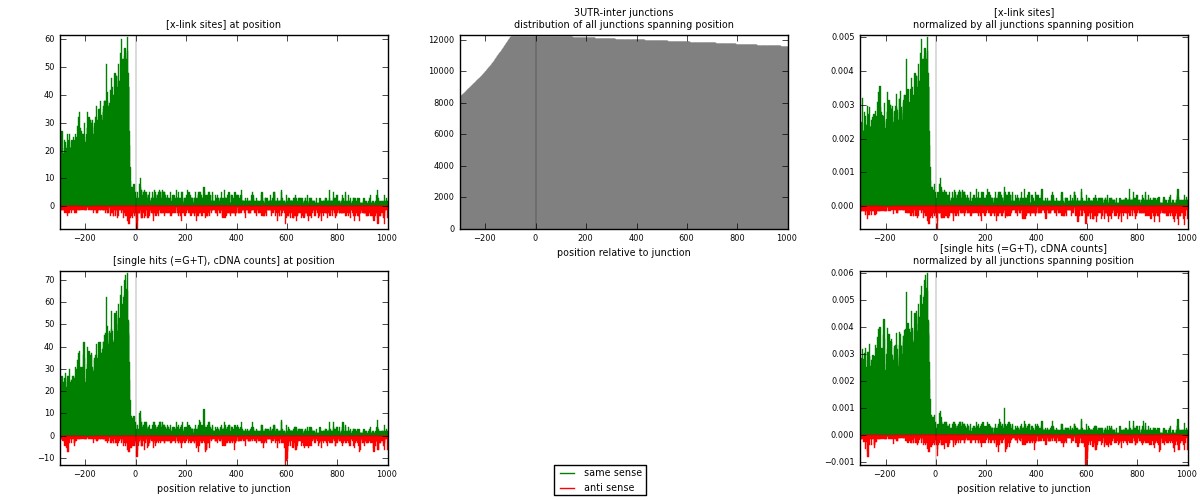
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10008 | 3.68% | 12.29% | 86.69% | 3266 | 2.71146 |
| anti | 1537 | 0.56% | 1.89% | 13.31% | 579 | 0.416418 |
| both | 11545 | 4.24% | 14.17% | 100.00% | 3691 | 3.12788 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 11304 | 3.79% | 12.42% | 86.31% | 3266 | 3.06258 |
| anti | 1793 | 0.60% | 1.97% | 13.69% | 579 | 0.485776 |
| both | 13097 | 4.40% | 14.40% | 100.00% | 3691 | 3.54836 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
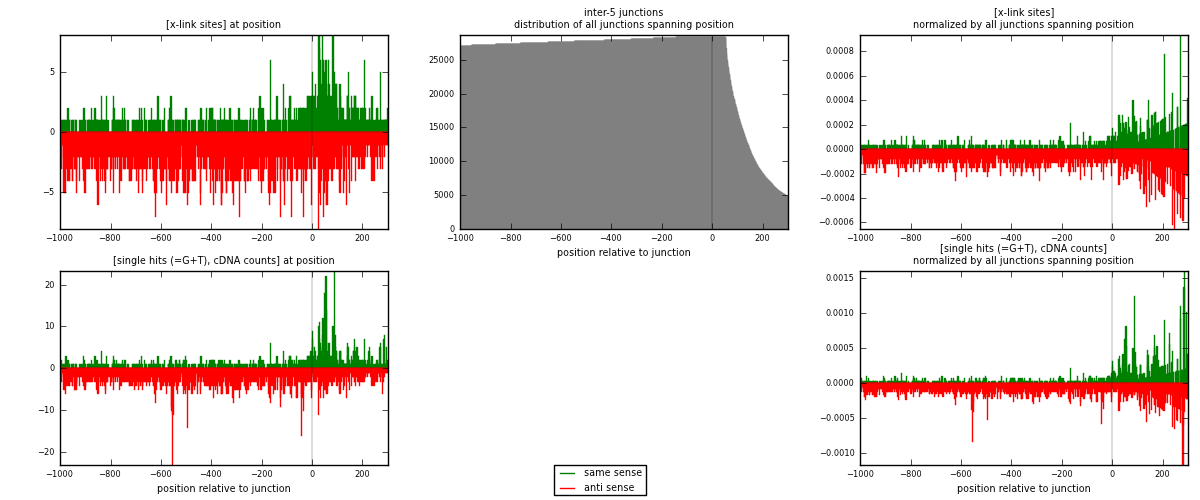
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 909 | 0.33% | 1.12% | 26.56% | 523 | 0.497265 |
| anti | 2514 | 0.92% | 3.09% | 73.44% | 1344 | 1.37527 |
| both | 3423 | 1.26% | 4.20% | 100.00% | 1828 | 1.87254 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1127 | 0.38% | 1.24% | 28.50% | 523 | 0.616521 |
| anti | 2828 | 0.95% | 3.11% | 71.50% | 1344 | 1.54705 |
| both | 3955 | 1.33% | 4.35% | 100.00% | 1828 | 2.16357 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
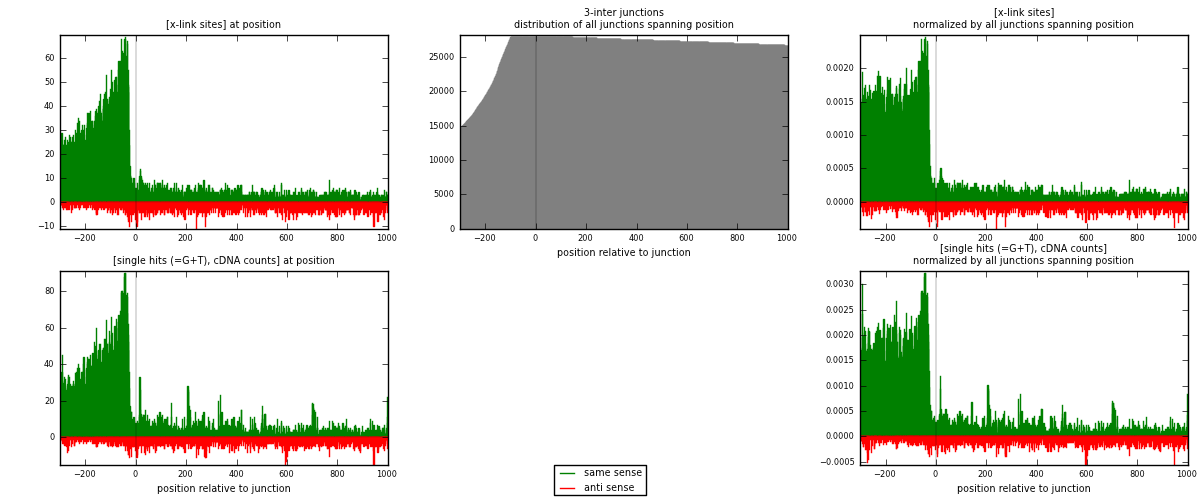
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12494 | 4.59% | 15.34% | 80.31% | 4061 | 2.4498 |
| anti | 3063 | 1.13% | 3.76% | 19.69% | 1238 | 0.600588 |
| both | 15557 | 5.72% | 19.10% | 100.00% | 5100 | 3.05039 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 15676 | 5.26% | 17.23% | 81.79% | 4061 | 3.07373 |
| anti | 3491 | 1.17% | 3.84% | 18.21% | 1238 | 0.68451 |
| both | 19167 | 6.43% | 21.07% | 100.00% | 5100 | 3.75824 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
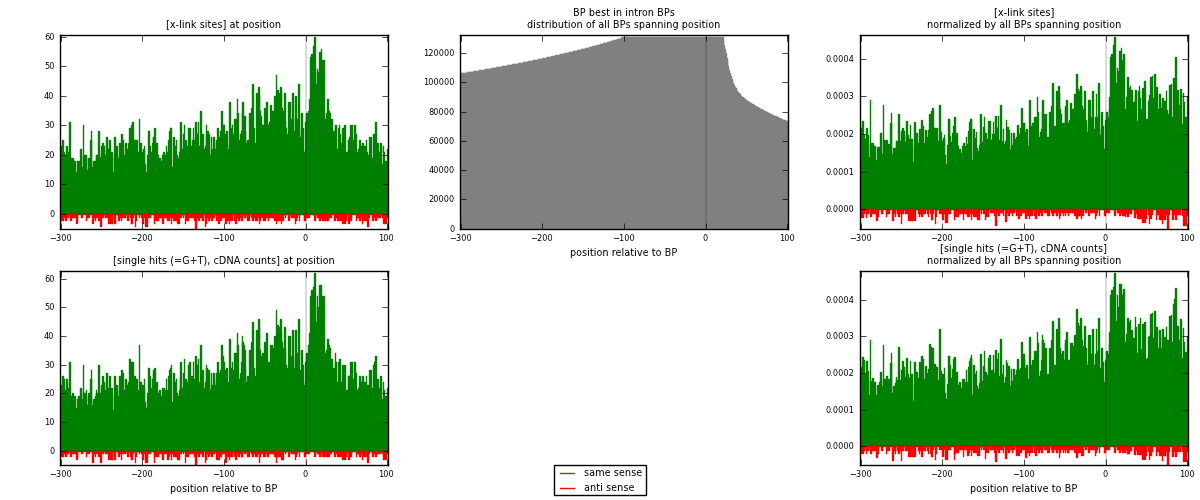
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 10306 | 3.79% | 12.65% | 95.95% | 8236 | 1.20173 |
| anti | 435 | 0.16% | 0.53% | 4.05% | 358 | 0.0507229 |
| both | 10741 | 3.95% | 13.19% | 100.00% | 8576 | 1.25245 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 10727 | 3.60% | 11.79% | 95.99% | 8236 | 1.25082 |
| anti | 448 | 0.15% | 0.49% | 4.01% | 358 | 0.0522388 |
| both | 11175 | 3.75% | 12.28% | 100.00% | 8576 | 1.30306 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.