RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
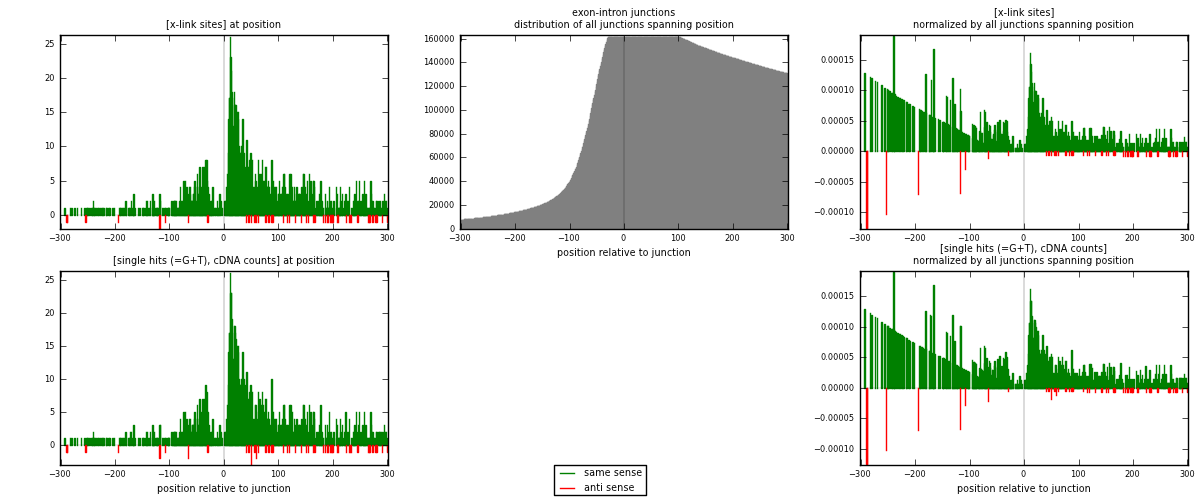 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1402 | 8.08% | 24.65% | 96.49% | 1306 | 1.0416 |
| anti | 51 | 0.29% | 0.90% | 3.51% | 40 | 0.03789 |
| both | 1453 | 8.37% | 25.55% | 100.00% | 1346 | 1.07949 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1443 | 7.84% | 23.95% | 96.33% | 1306 | 1.07207 |
| anti | 55 | 0.30% | 0.91% | 3.67% | 40 | 0.0408618 |
| both | 1498 | 8.14% | 24.86% | 100.00% | 1346 | 1.11293 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
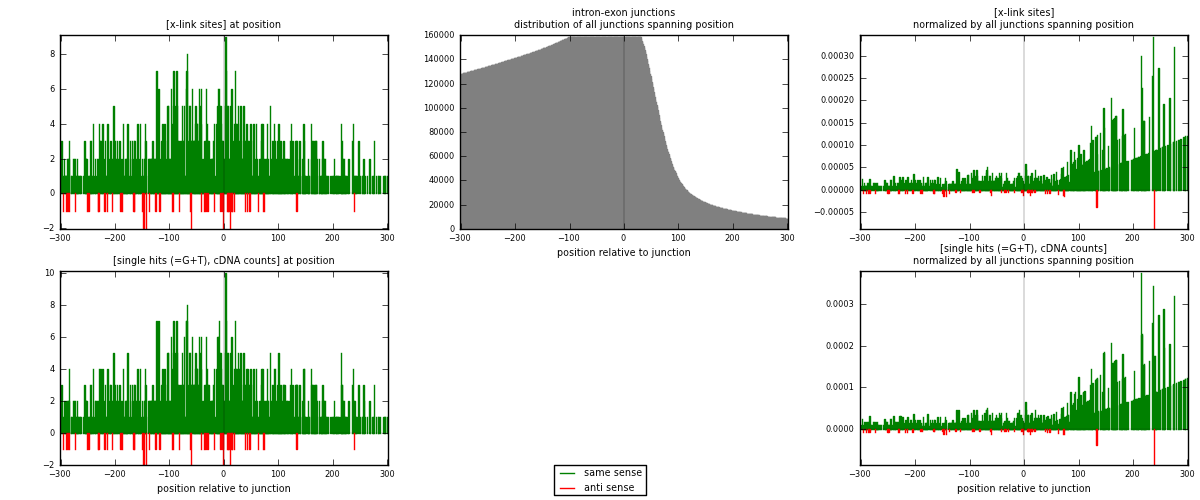 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1074 | 6.19% | 18.88% | 95.13% | 970 | 1.05191 |
| anti | 55 | 0.32% | 0.97% | 4.87% | 51 | 0.0538688 |
| both | 1129 | 6.50% | 19.85% | 100.00% | 1021 | 1.10578 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1106 | 6.01% | 18.36% | 95.26% | 970 | 1.08325 |
| anti | 55 | 0.30% | 0.91% | 4.74% | 51 | 0.0538688 |
| both | 1161 | 6.31% | 19.27% | 100.00% | 1021 | 1.13712 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
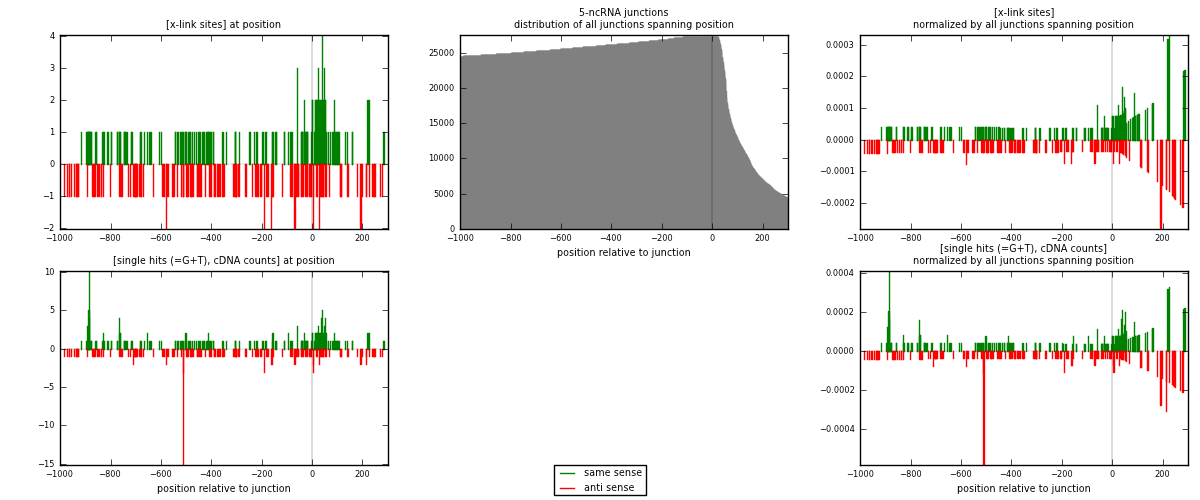 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 199 | 1.15% | 3.50% | 54.82% | 99 | 0.917051 |
| anti | 164 | 0.94% | 2.88% | 45.18% | 119 | 0.75576 |
| both | 363 | 2.09% | 6.38% | 100.00% | 217 | 1.67281 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 237 | 1.29% | 3.93% | 56.03% | 99 | 1.09217 |
| anti | 186 | 1.01% | 3.09% | 43.97% | 119 | 0.857143 |
| both | 423 | 2.30% | 7.02% | 100.00% | 217 | 1.94931 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
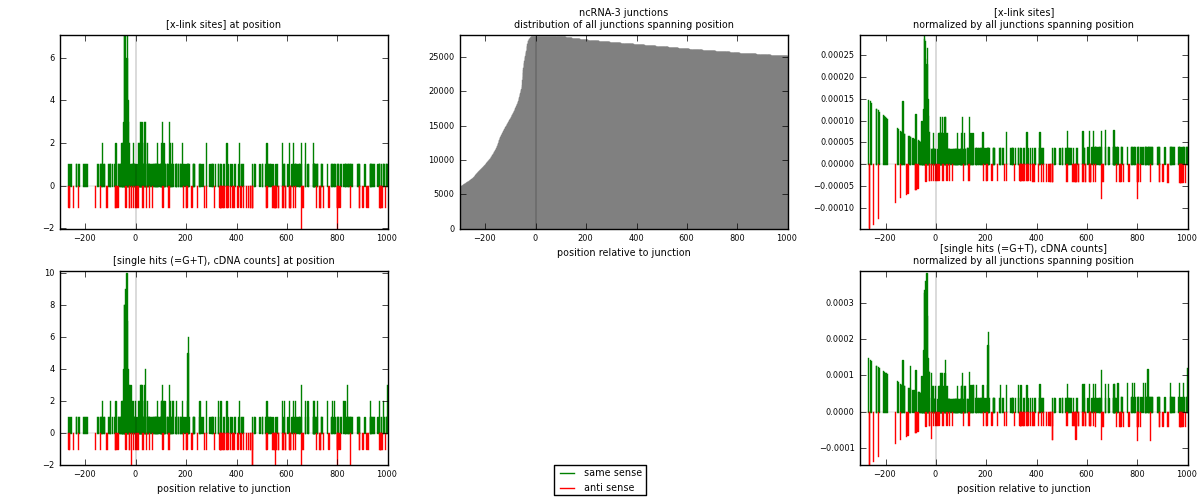 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 401 | 2.31% | 7.05% | 78.17% | 224 | 1.30619 |
| anti | 112 | 0.65% | 1.97% | 21.83% | 83 | 0.364821 |
| both | 513 | 2.96% | 9.02% | 100.00% | 307 | 1.67101 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 458 | 2.49% | 7.60% | 79.65% | 224 | 1.49186 |
| anti | 117 | 0.64% | 1.94% | 20.35% | 83 | 0.381107 |
| both | 575 | 3.12% | 9.54% | 100.00% | 307 | 1.87296 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
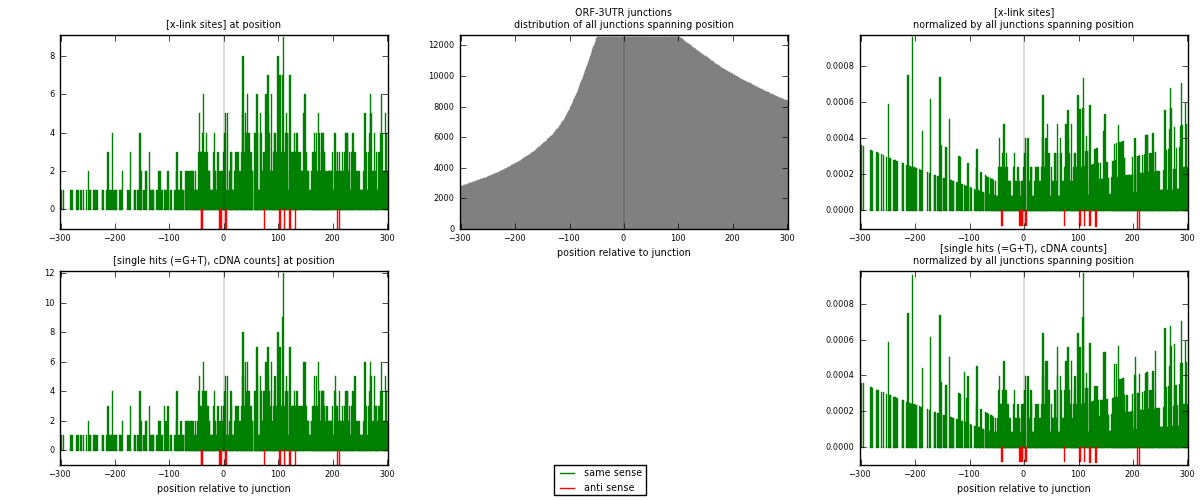 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 888 | 5.12% | 15.61% | 98.67% | 572 | 1.52055 |
| anti | 12 | 0.07% | 0.21% | 1.33% | 12 | 0.0205479 |
| both | 900 | 5.19% | 15.82% | 100.00% | 584 | 1.5411 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 939 | 5.10% | 15.59% | 98.74% | 572 | 1.60788 |
| anti | 12 | 0.07% | 0.20% | 1.26% | 12 | 0.0205479 |
| both | 951 | 5.17% | 15.78% | 100.00% | 584 | 1.62842 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
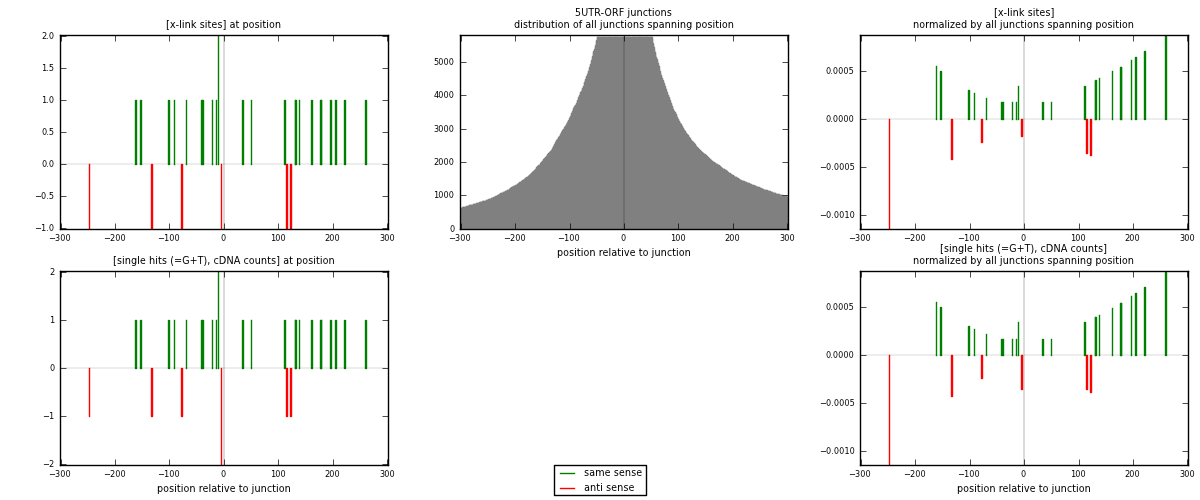 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 24 | 0.14% | 0.42% | 80.00% | 23 | 0.857143 |
| anti | 6 | 0.03% | 0.11% | 20.00% | 5 | 0.214286 |
| both | 30 | 0.17% | 0.53% | 100.00% | 28 | 1.07143 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 24 | 0.13% | 0.40% | 77.42% | 23 | 0.857143 |
| anti | 7 | 0.04% | 0.12% | 22.58% | 5 | 0.25 |
| both | 31 | 0.17% | 0.51% | 100.00% | 28 | 1.10714 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
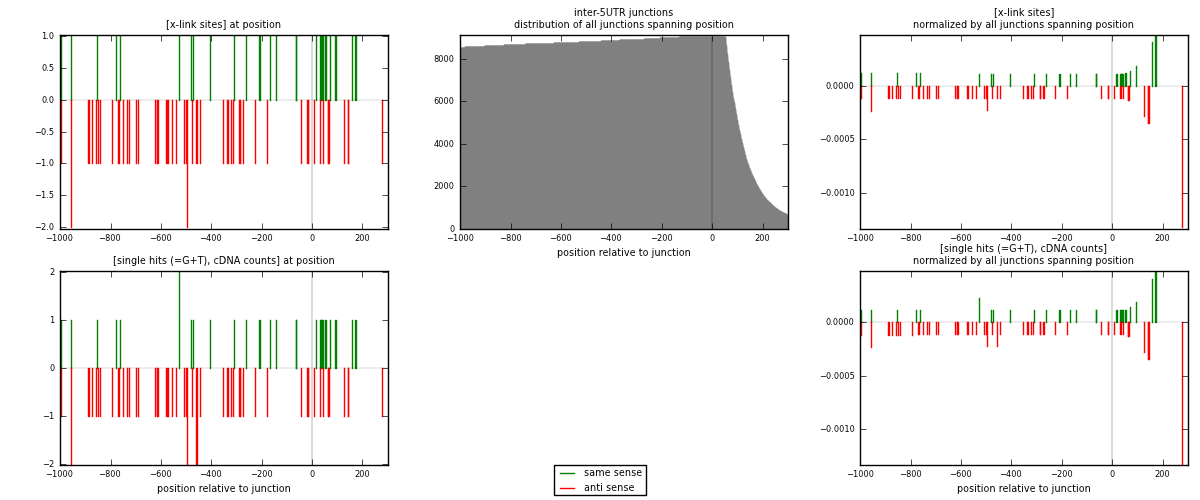 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 26 | 0.15% | 0.46% | 33.33% | 23 | 0.38806 |
| anti | 52 | 0.30% | 0.91% | 66.67% | 44 | 0.776119 |
| both | 78 | 0.45% | 1.37% | 100.00% | 67 | 1.16418 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 27 | 0.15% | 0.45% | 33.75% | 23 | 0.402985 |
| anti | 53 | 0.29% | 0.88% | 66.25% | 44 | 0.791045 |
| both | 80 | 0.43% | 1.33% | 100.00% | 67 | 1.19403 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
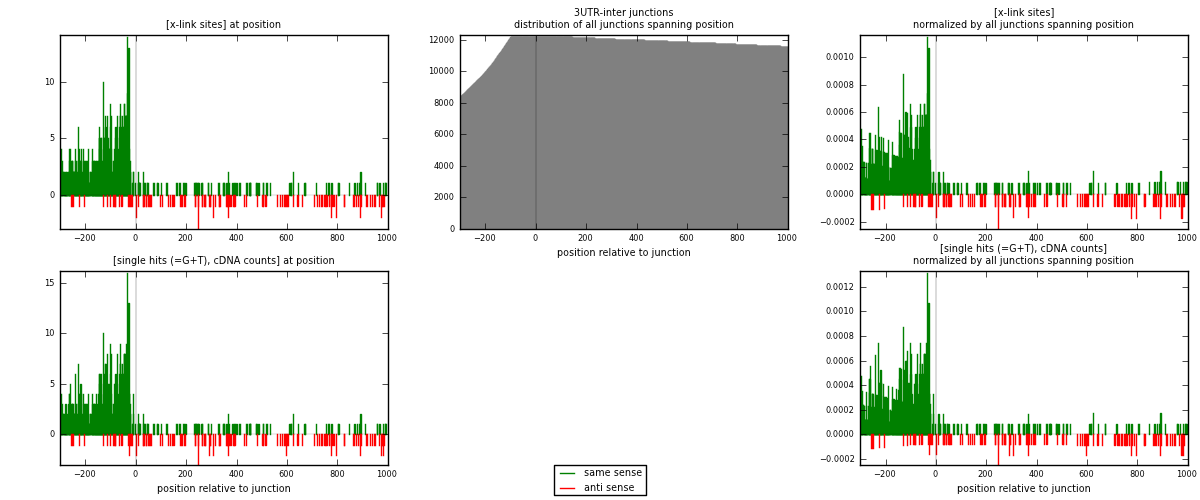 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 890 | 5.13% | 15.65% | 87.43% | 611 | 1.28242 |
| anti | 128 | 0.74% | 2.25% | 12.57% | 88 | 0.184438 |
| both | 1018 | 5.87% | 17.90% | 100.00% | 694 | 1.46686 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 941 | 5.11% | 15.62% | 87.70% | 611 | 1.35591 |
| anti | 132 | 0.72% | 2.19% | 12.30% | 88 | 0.190202 |
| both | 1073 | 5.83% | 17.81% | 100.00% | 694 | 1.54611 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
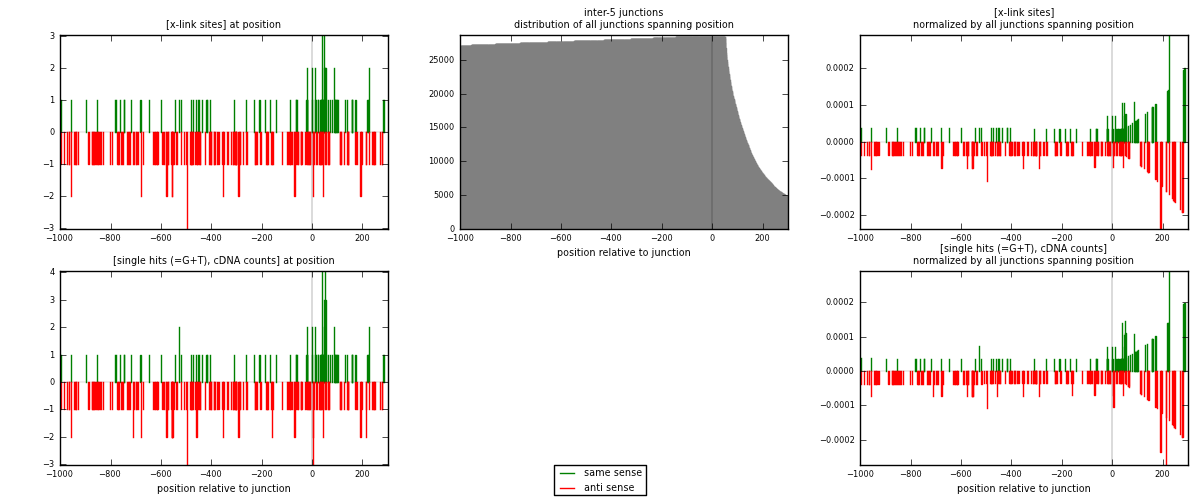
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 95 | 0.55% | 1.67% | 32.20% | 69 | 0.446009 |
| anti | 200 | 1.15% | 3.52% | 67.80% | 145 | 0.938967 |
| both | 295 | 1.70% | 5.19% | 100.00% | 213 | 1.38498 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 100 | 0.54% | 1.66% | 32.47% | 69 | 0.469484 |
| anti | 208 | 1.13% | 3.45% | 67.53% | 145 | 0.976526 |
| both | 308 | 1.67% | 5.11% | 100.00% | 213 | 1.44601 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
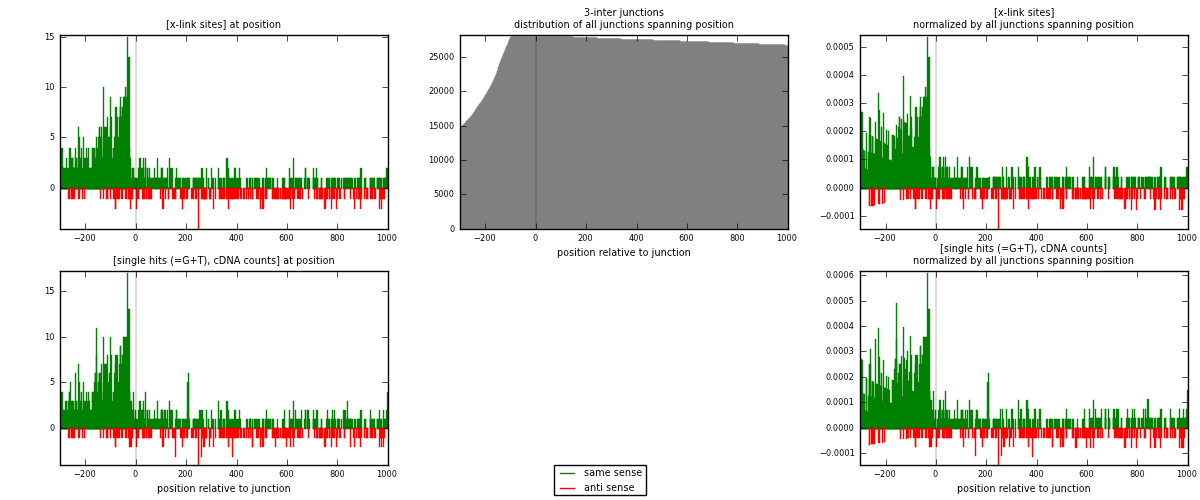
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1225 | 7.06% | 21.54% | 82.99% | 761 | 1.34615 |
| anti | 251 | 1.45% | 4.41% | 17.01% | 155 | 0.275824 |
| both | 1476 | 8.50% | 25.95% | 100.00% | 910 | 1.62198 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1343 | 7.30% | 22.29% | 83.36% | 761 | 1.47582 |
| anti | 268 | 1.46% | 4.45% | 16.64% | 155 | 0.294505 |
| both | 1611 | 8.75% | 26.74% | 100.00% | 910 | 1.77033 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
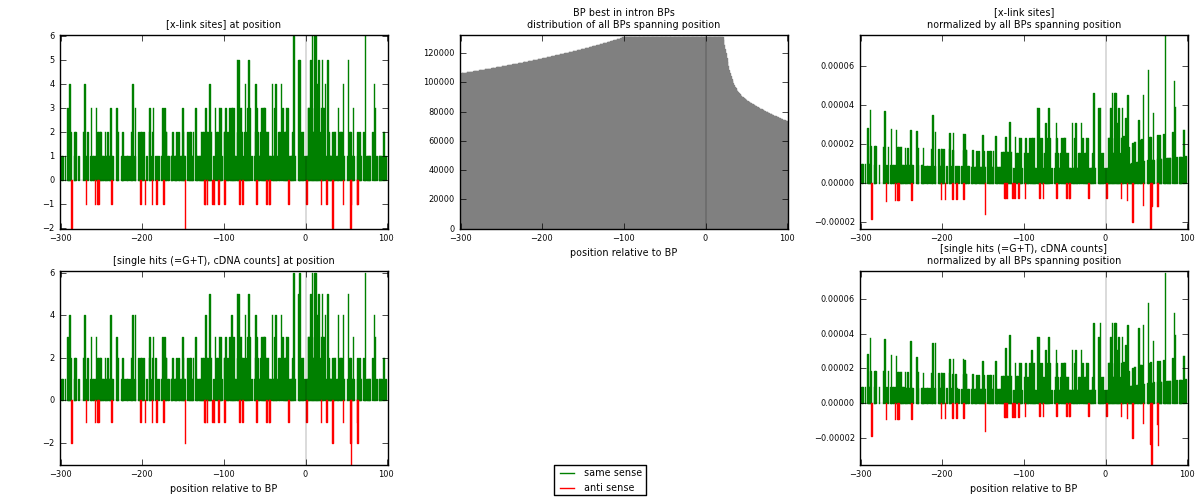
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 558 | 3.21% | 9.81% | 93.47% | 522 | 0.996429 |
| anti | 39 | 0.22% | 0.69% | 6.53% | 38 | 0.0696429 |
| both | 597 | 3.44% | 10.50% | 100.00% | 560 | 1.06607 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 567 | 3.08% | 9.41% | 93.10% | 522 | 1.0125 |
| anti | 42 | 0.23% | 0.70% | 6.90% | 38 | 0.075 |
| both | 609 | 3.31% | 10.11% | 100.00% | 560 | 1.0875 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.