RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
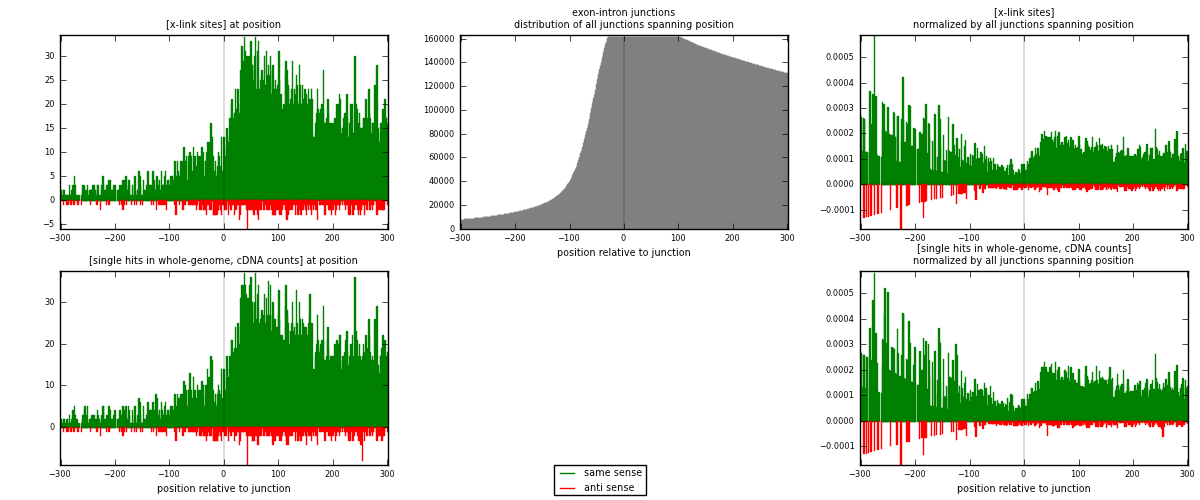 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6568 | 3.97% | 28.86% | 93.22% | 4500 | 1.3385 |
| anti | 478 | 0.29% | 2.10% | 6.78% | 417 | 0.0974119 |
| both | 7046 | 4.26% | 30.96% | 100.00% | 4907 | 1.43591 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 7048 | 3.87% | 24.62% | 93.35% | 4500 | 1.43632 |
| anti | 502 | 0.28% | 1.75% | 6.65% | 417 | 0.102303 |
| both | 7550 | 4.15% | 26.38% | 100.00% | 4907 | 1.53862 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
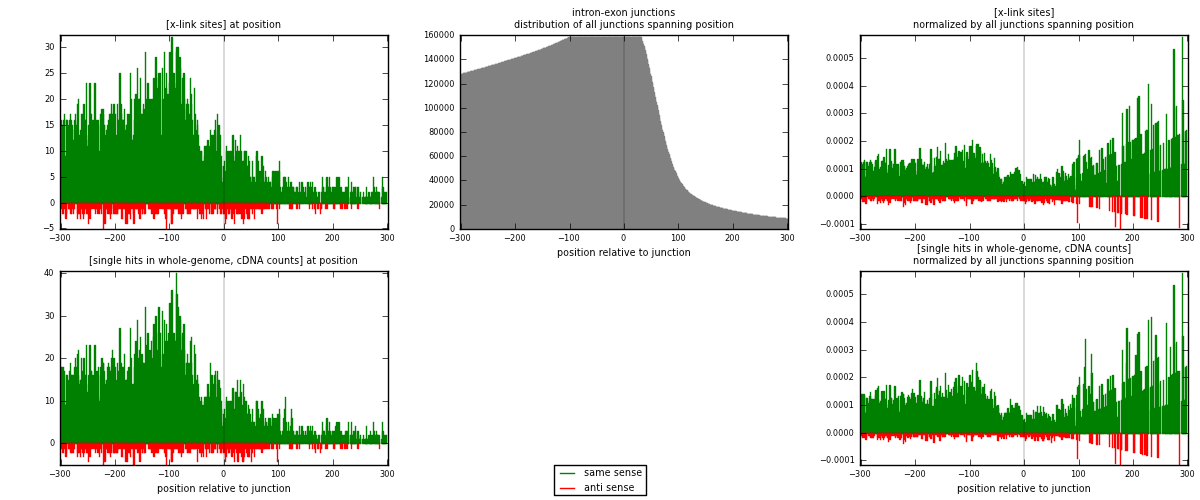 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5625 | 3.40% | 24.72% | 92.08% | 4074 | 1.25614 |
| anti | 484 | 0.29% | 2.13% | 7.92% | 416 | 0.108084 |
| both | 6109 | 3.69% | 26.85% | 100.00% | 4478 | 1.36423 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 6068 | 3.33% | 21.20% | 92.42% | 4074 | 1.35507 |
| anti | 498 | 0.27% | 1.74% | 7.58% | 416 | 0.11121 |
| both | 6566 | 3.61% | 22.94% | 100.00% | 4478 | 1.46628 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
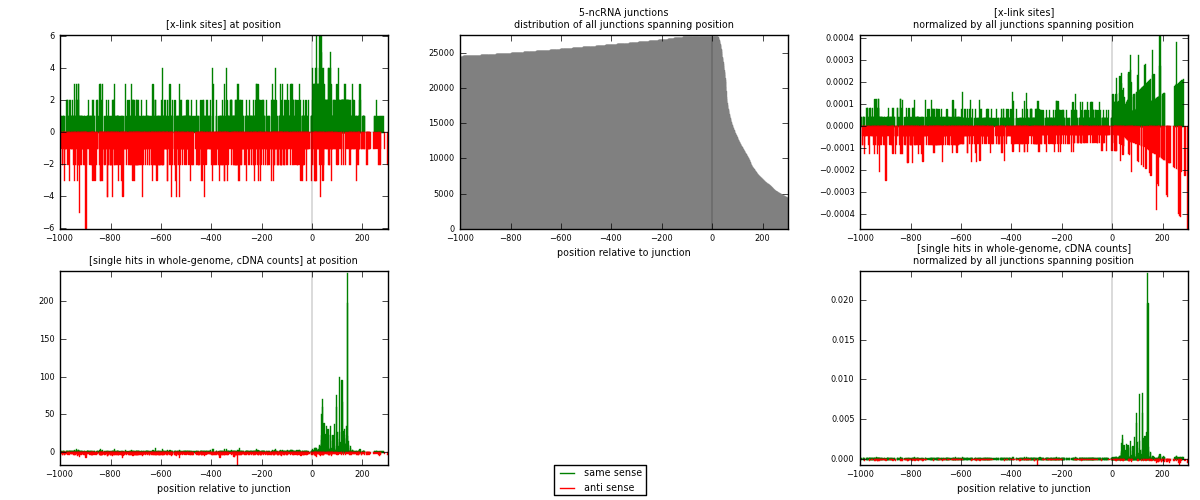 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1005 | 0.61% | 4.42% | 48.53% | 507 | 0.851695 |
| anti | 1066 | 0.64% | 4.68% | 51.47% | 681 | 0.90339 |
| both | 2071 | 1.25% | 9.10% | 100.00% | 1180 | 1.75508 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2857 | 1.57% | 9.98% | 71.03% | 507 | 2.42119 |
| anti | 1165 | 0.64% | 4.07% | 28.97% | 681 | 0.987288 |
| both | 4022 | 2.21% | 14.05% | 100.00% | 1180 | 3.40847 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
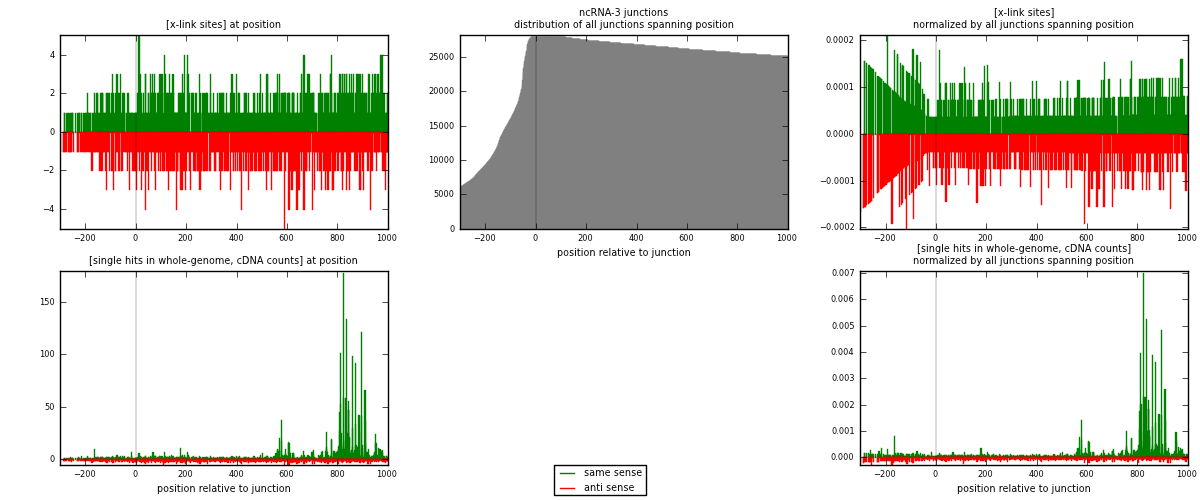 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1115 | 0.67% | 4.90% | 53.27% | 500 | 1.0018 |
| anti | 978 | 0.59% | 4.30% | 46.73% | 621 | 0.878706 |
| both | 2093 | 1.26% | 9.20% | 100.00% | 1113 | 1.8805 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3519 | 1.93% | 12.29% | 76.95% | 500 | 3.16173 |
| anti | 1054 | 0.58% | 3.68% | 23.05% | 621 | 0.94699 |
| both | 4573 | 2.51% | 15.98% | 100.00% | 1113 | 4.10872 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
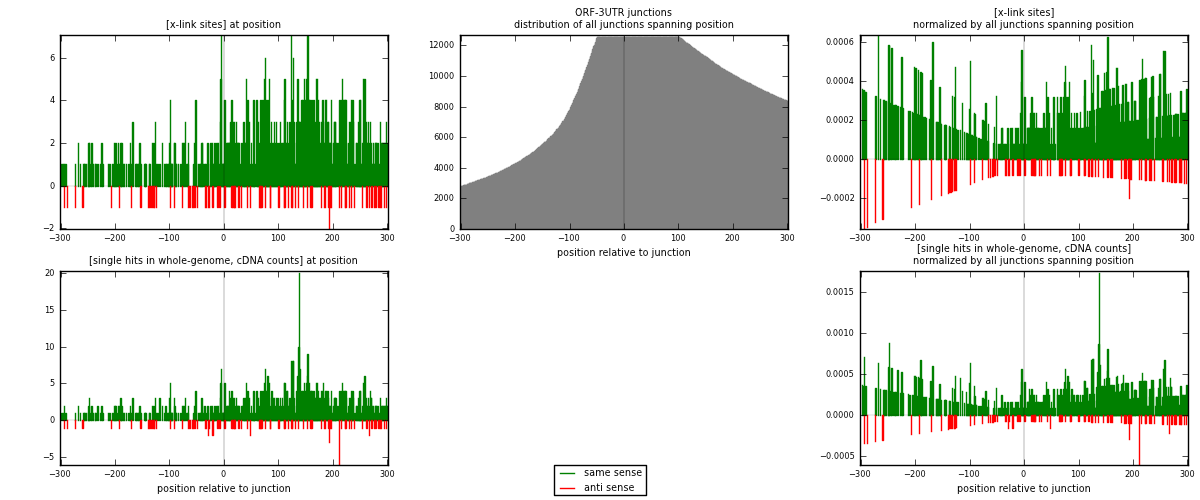
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 790 | 0.48% | 3.47% | 90.18% | 493 | 1.3884 |
| anti | 86 | 0.05% | 0.38% | 9.82% | 79 | 0.151142 |
| both | 876 | 0.53% | 3.85% | 100.00% | 569 | 1.53954 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 894 | 0.49% | 3.12% | 90.30% | 493 | 1.57118 |
| anti | 96 | 0.05% | 0.34% | 9.70% | 79 | 0.168717 |
| both | 990 | 0.54% | 3.46% | 100.00% | 569 | 1.73989 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
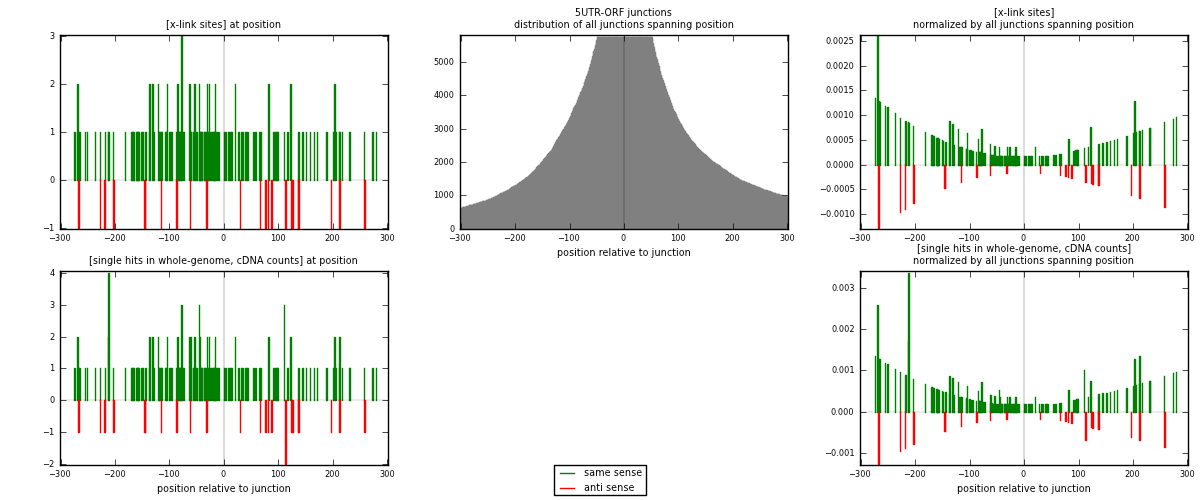
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 142 | 0.09% | 0.62% | 86.06% | 88 | 1.33962 |
| anti | 23 | 0.01% | 0.10% | 13.94% | 18 | 0.216981 |
| both | 165 | 0.10% | 0.73% | 100.00% | 106 | 1.5566 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 152 | 0.08% | 0.53% | 86.36% | 88 | 1.43396 |
| anti | 24 | 0.01% | 0.08% | 13.64% | 18 | 0.226415 |
| both | 176 | 0.10% | 0.61% | 100.00% | 106 | 1.66038 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
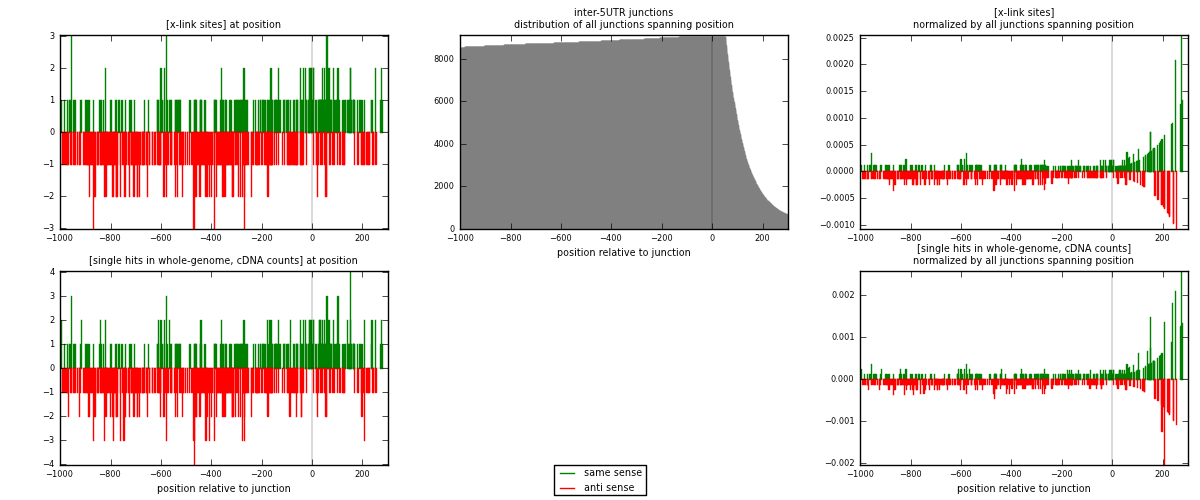
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 260 | 0.16% | 1.14% | 40.75% | 179 | 0.625 |
| anti | 378 | 0.23% | 1.66% | 59.25% | 240 | 0.908654 |
| both | 638 | 0.39% | 2.80% | 100.00% | 416 | 1.53365 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 278 | 0.15% | 0.97% | 40.64% | 179 | 0.668269 |
| anti | 406 | 0.22% | 1.42% | 59.36% | 240 | 0.975962 |
| both | 684 | 0.38% | 2.39% | 100.00% | 416 | 1.64423 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
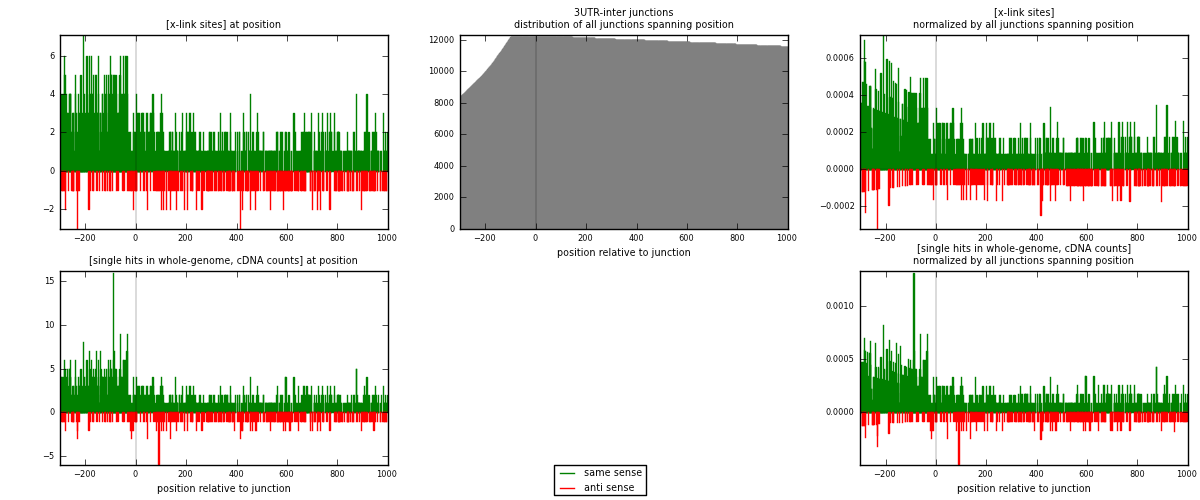
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1304 | 0.79% | 5.73% | 80.59% | 824 | 1.27468 |
| anti | 314 | 0.19% | 1.38% | 19.41% | 213 | 0.30694 |
| both | 1618 | 0.98% | 7.11% | 100.00% | 1023 | 1.58162 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1417 | 0.78% | 4.95% | 80.74% | 824 | 1.38514 |
| anti | 338 | 0.19% | 1.18% | 19.26% | 213 | 0.330401 |
| both | 1755 | 0.96% | 6.13% | 100.00% | 1023 | 1.71554 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
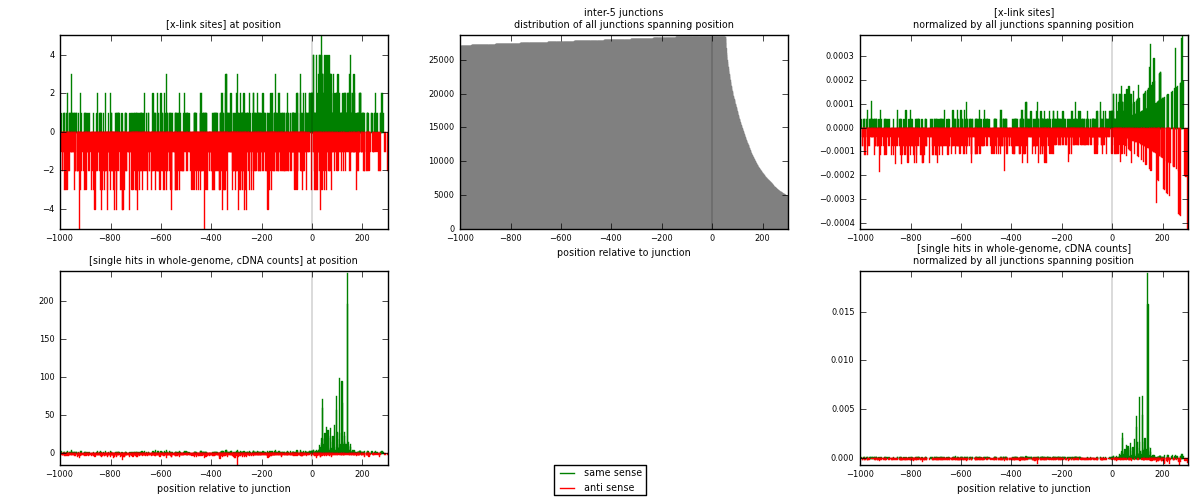 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 637 | 0.38% | 2.80% | 34.01% | 367 | 0.557305 |
| anti | 1236 | 0.75% | 5.43% | 65.99% | 784 | 1.08136 |
| both | 1873 | 1.13% | 8.23% | 100.00% | 1143 | 1.63867 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2276 | 1.25% | 7.95% | 62.73% | 367 | 1.99125 |
| anti | 1352 | 0.74% | 4.72% | 37.27% | 784 | 1.18285 |
| both | 3628 | 1.99% | 12.68% | 100.00% | 1143 | 3.1741 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
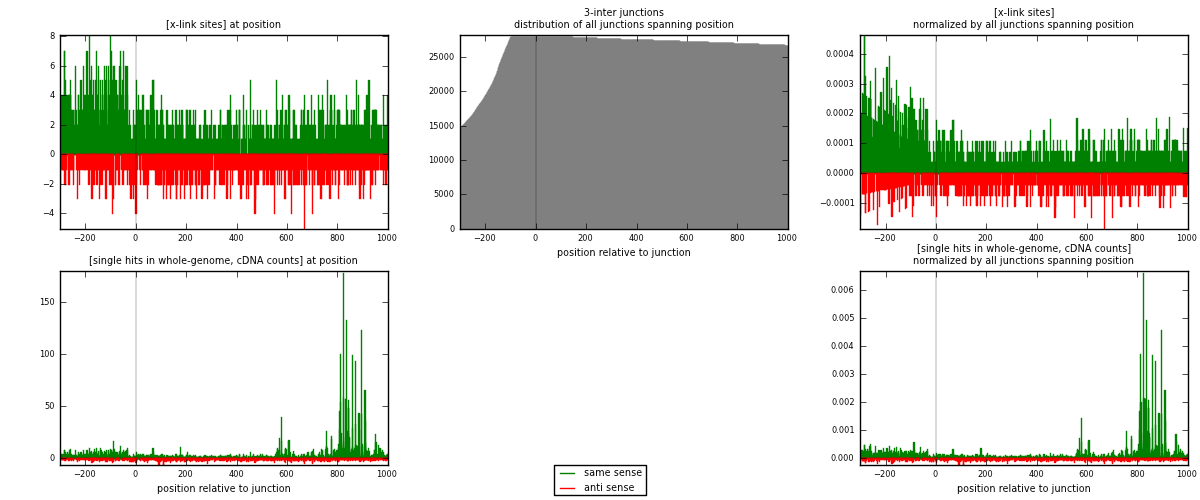 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1978 | 1.19% | 8.69% | 66.91% | 1025 | 1.21053 |
| anti | 978 | 0.59% | 4.30% | 33.09% | 625 | 0.598531 |
| both | 2956 | 1.79% | 12.99% | 100.00% | 1634 | 1.80906 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4460 | 2.45% | 15.58% | 80.87% | 1025 | 2.7295 |
| anti | 1055 | 0.58% | 3.69% | 19.13% | 625 | 0.645655 |
| both | 5515 | 3.03% | 19.27% | 100.00% | 1634 | 3.37515 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
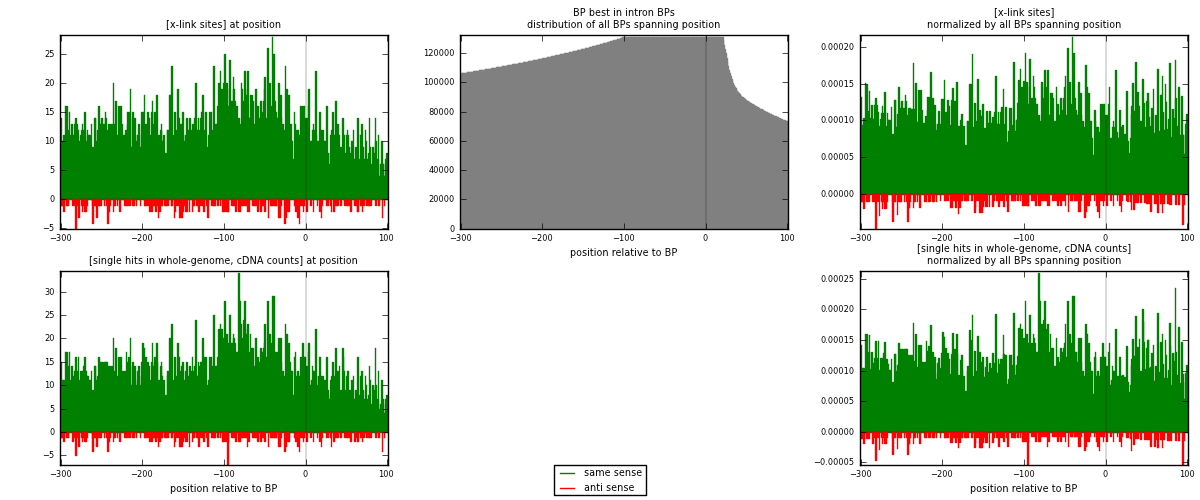 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 4815 | 2.91% | 21.16% | 94.13% | 3353 | 1.33639 |
| anti | 300 | 0.18% | 1.32% | 5.87% | 255 | 0.0832639 |
| both | 5115 | 3.09% | 22.48% | 100.00% | 3603 | 1.41965 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 5217 | 2.87% | 18.23% | 94.34% | 3353 | 1.44796 |
| anti | 313 | 0.17% | 1.09% | 5.66% | 255 | 0.0868721 |
| both | 5530 | 3.04% | 19.32% | 100.00% | 3603 | 1.53483 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.