RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
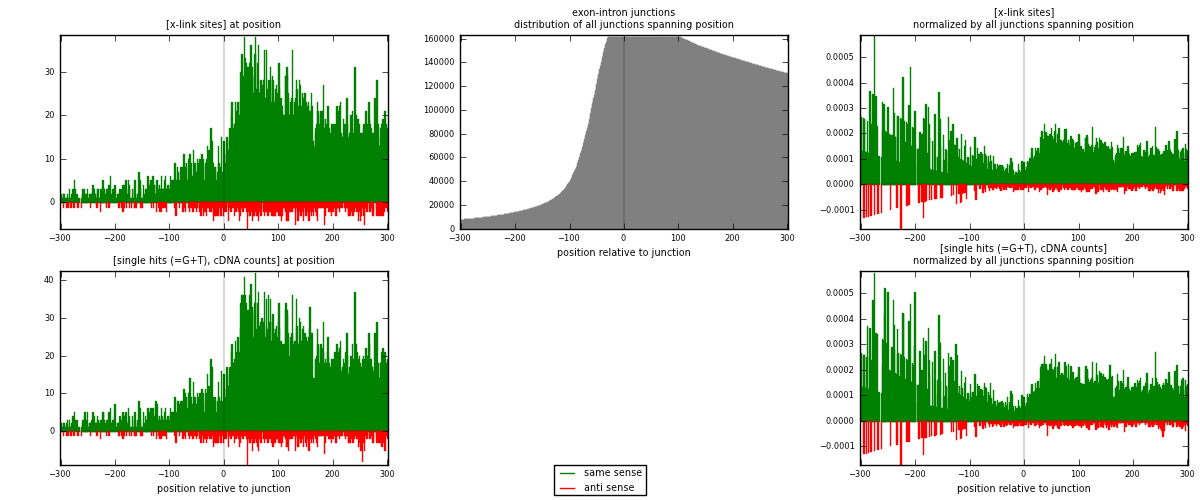 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7004 | 3.85% | 28.40% | 92.27% | 4788 | 1.32326 |
| anti | 587 | 0.32% | 2.38% | 7.73% | 520 | 0.110901 |
| both | 7591 | 4.18% | 30.78% | 100.00% | 5293 | 1.43416 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 7515 | 3.77% | 24.46% | 92.41% | 4788 | 1.4198 |
| anti | 617 | 0.31% | 2.01% | 7.59% | 520 | 0.116569 |
| both | 8132 | 4.07% | 26.47% | 100.00% | 5293 | 1.53637 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
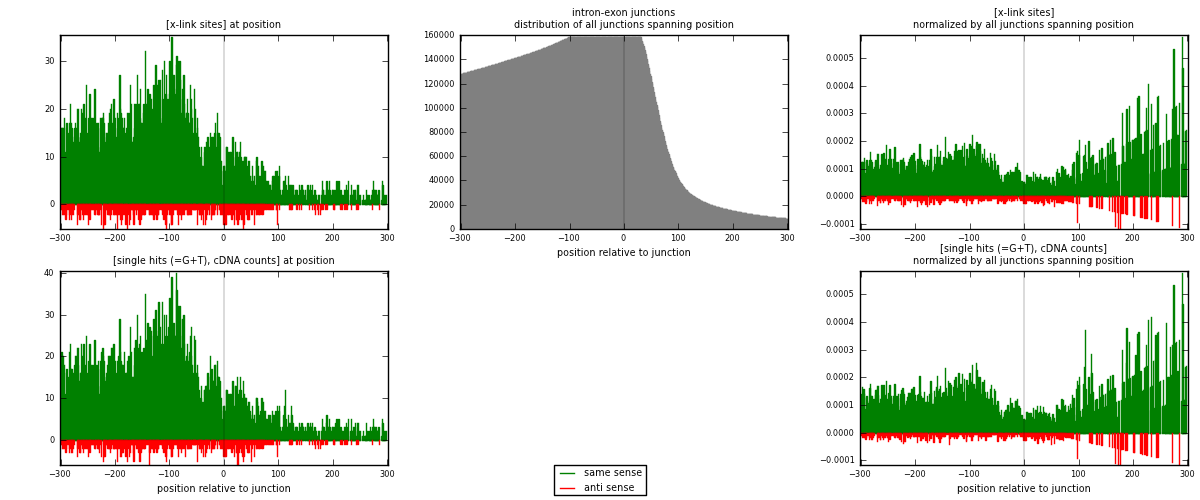 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6016 | 3.31% | 24.39% | 90.68% | 4353 | 1.2343 |
| anti | 618 | 0.34% | 2.51% | 9.32% | 539 | 0.126795 |
| both | 6634 | 3.65% | 26.90% | 100.00% | 4874 | 1.3611 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 6491 | 3.25% | 21.13% | 91.00% | 4353 | 1.33176 |
| anti | 642 | 0.32% | 2.09% | 9.00% | 539 | 0.131719 |
| both | 7133 | 3.57% | 23.22% | 100.00% | 4874 | 1.46348 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
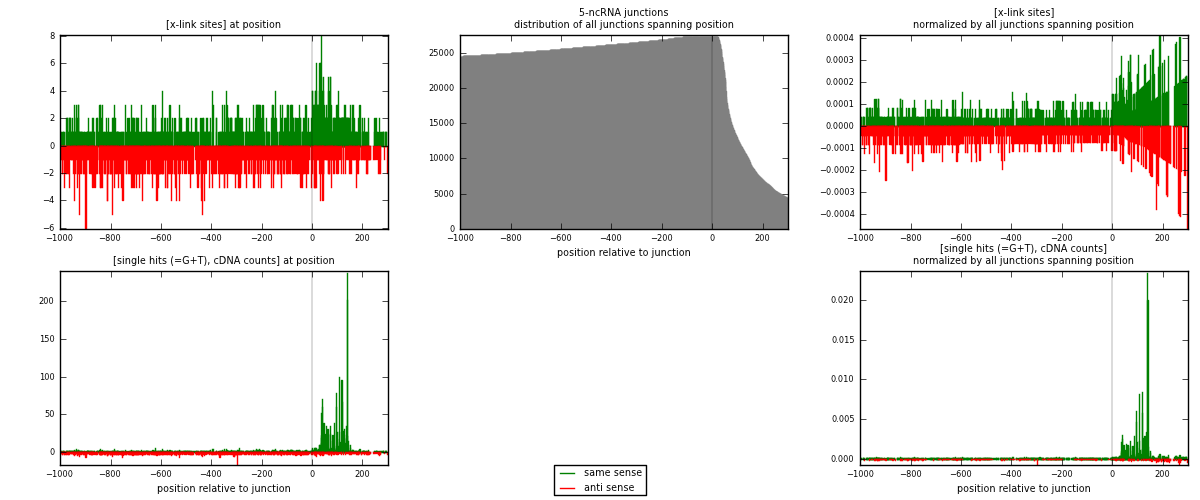 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1112 | 0.61% | 4.51% | 48.81% | 562 | 0.857363 |
| anti | 1166 | 0.64% | 4.73% | 51.19% | 748 | 0.898998 |
| both | 2278 | 1.25% | 9.24% | 100.00% | 1297 | 1.75636 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3011 | 1.51% | 9.80% | 70.30% | 562 | 2.32151 |
| anti | 1272 | 0.64% | 4.14% | 29.70% | 748 | 0.980725 |
| both | 4283 | 2.15% | 13.94% | 100.00% | 1297 | 3.30224 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
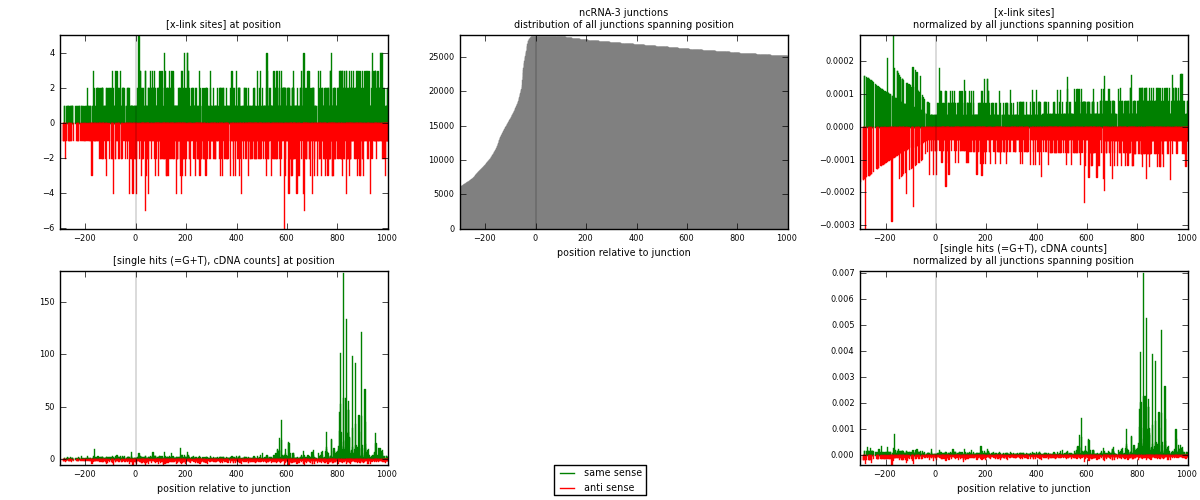 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1200 | 0.66% | 4.86% | 52.17% | 559 | 0.967742 |
| anti | 1100 | 0.61% | 4.46% | 47.83% | 690 | 0.887097 |
| both | 2300 | 1.27% | 9.32% | 100.00% | 1240 | 1.85484 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3612 | 1.81% | 11.76% | 75.33% | 559 | 2.9129 |
| anti | 1183 | 0.59% | 3.85% | 24.67% | 690 | 0.954032 |
| both | 4795 | 2.40% | 15.61% | 100.00% | 1240 | 3.86694 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
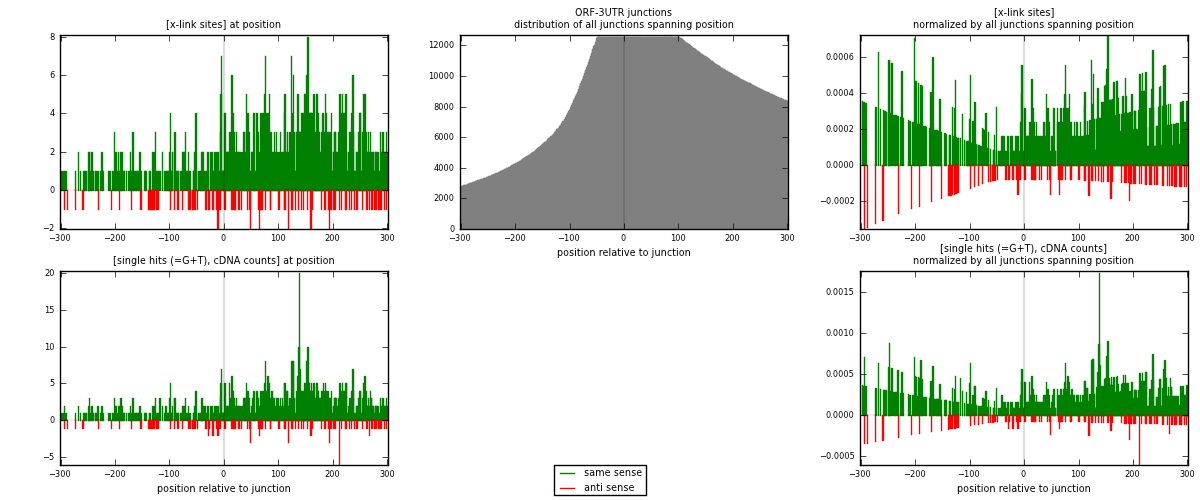
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 845 | 0.46% | 3.43% | 89.23% | 523 | 1.38072 |
| anti | 102 | 0.06% | 0.41% | 10.77% | 93 | 0.166667 |
| both | 947 | 0.52% | 3.84% | 100.00% | 612 | 1.54739 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 958 | 0.48% | 3.12% | 89.45% | 523 | 1.56536 |
| anti | 113 | 0.06% | 0.37% | 10.55% | 93 | 0.184641 |
| both | 1071 | 0.54% | 3.49% | 100.00% | 612 | 1.75 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
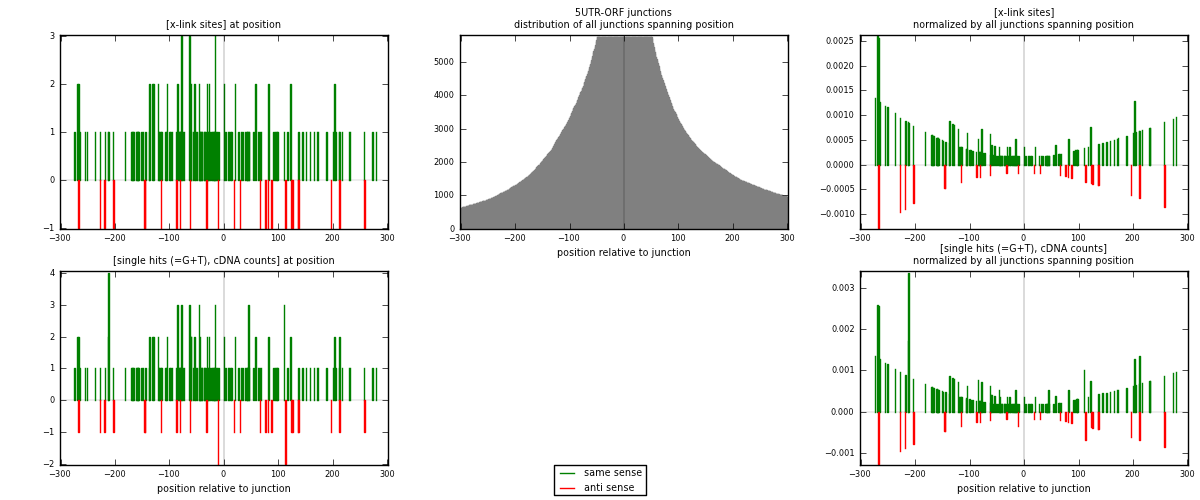
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 155 | 0.09% | 0.63% | 85.64% | 98 | 1.30252 |
| anti | 26 | 0.01% | 0.11% | 14.36% | 21 | 0.218487 |
| both | 181 | 0.10% | 0.73% | 100.00% | 119 | 1.52101 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 168 | 0.08% | 0.55% | 85.71% | 98 | 1.41176 |
| anti | 28 | 0.01% | 0.09% | 14.29% | 21 | 0.235294 |
| both | 196 | 0.10% | 0.64% | 100.00% | 119 | 1.64706 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
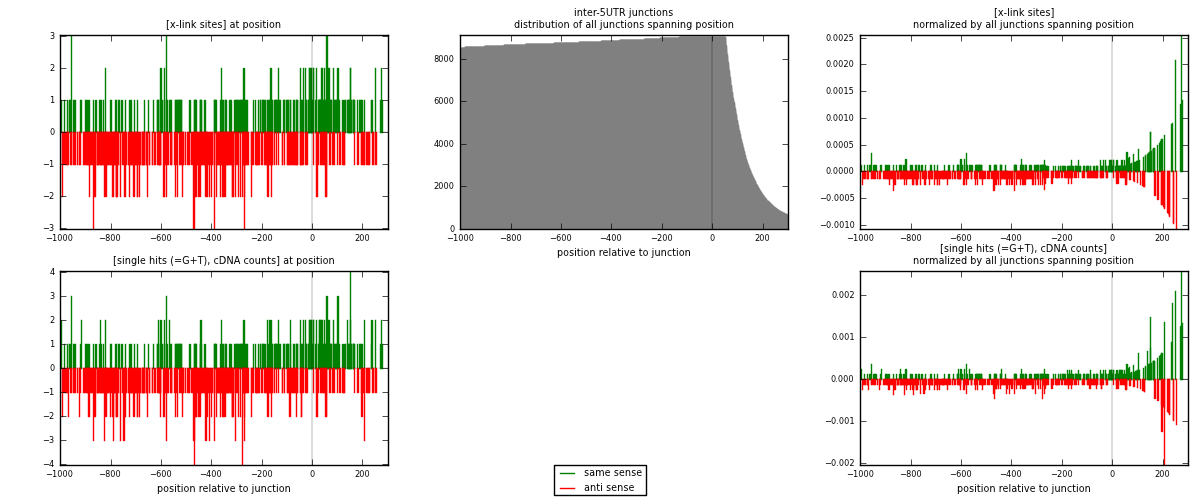
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 280 | 0.15% | 1.14% | 41.12% | 192 | 0.625 |
| anti | 401 | 0.22% | 1.63% | 58.88% | 259 | 0.895089 |
| both | 681 | 0.37% | 2.76% | 100.00% | 448 | 1.52009 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 299 | 0.15% | 0.97% | 41.02% | 192 | 0.667411 |
| anti | 430 | 0.22% | 1.40% | 58.98% | 259 | 0.959821 |
| both | 729 | 0.37% | 2.37% | 100.00% | 448 | 1.62723 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
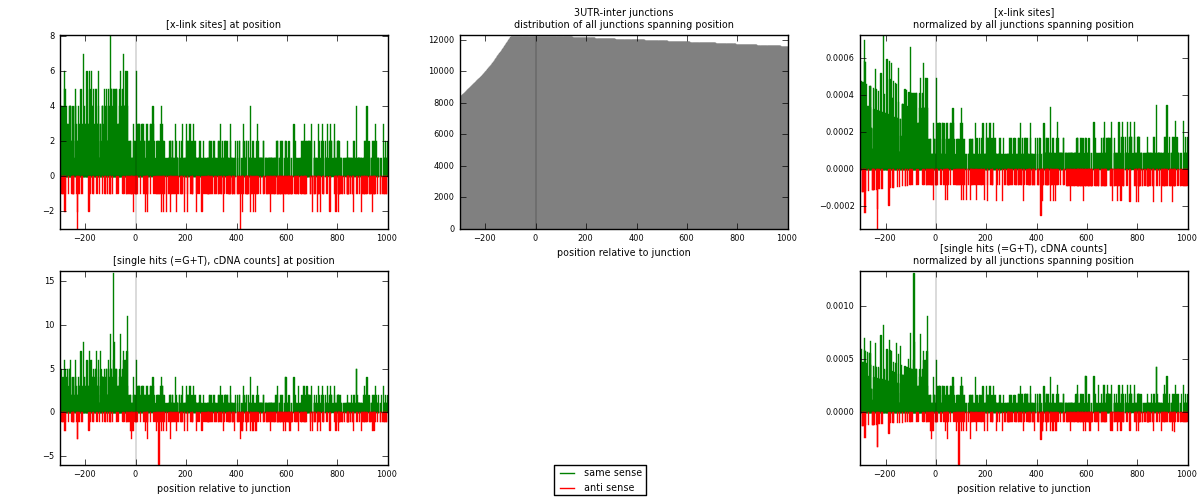
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1365 | 0.75% | 5.53% | 80.01% | 861 | 1.26623 |
| anti | 341 | 0.19% | 1.38% | 19.99% | 232 | 0.316327 |
| both | 1706 | 0.94% | 6.92% | 100.00% | 1078 | 1.58256 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1486 | 0.74% | 4.84% | 80.24% | 861 | 1.37848 |
| anti | 366 | 0.18% | 1.19% | 19.76% | 232 | 0.339518 |
| both | 1852 | 0.93% | 6.03% | 100.00% | 1078 | 1.718 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
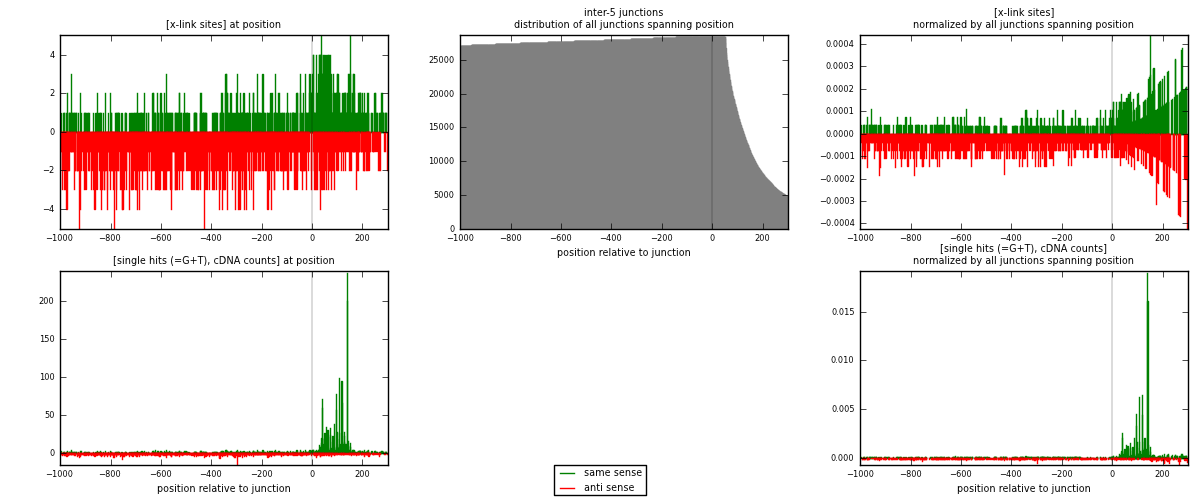
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 723 | 0.40% | 2.93% | 35.06% | 413 | 0.575179 |
| anti | 1339 | 0.74% | 5.43% | 64.94% | 855 | 1.06523 |
| both | 2062 | 1.13% | 8.36% | 100.00% | 1257 | 1.64041 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2391 | 1.20% | 7.78% | 62.12% | 413 | 1.90215 |
| anti | 1458 | 0.73% | 4.75% | 37.88% | 855 | 1.1599 |
| both | 3849 | 1.93% | 12.53% | 100.00% | 1257 | 3.06205 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
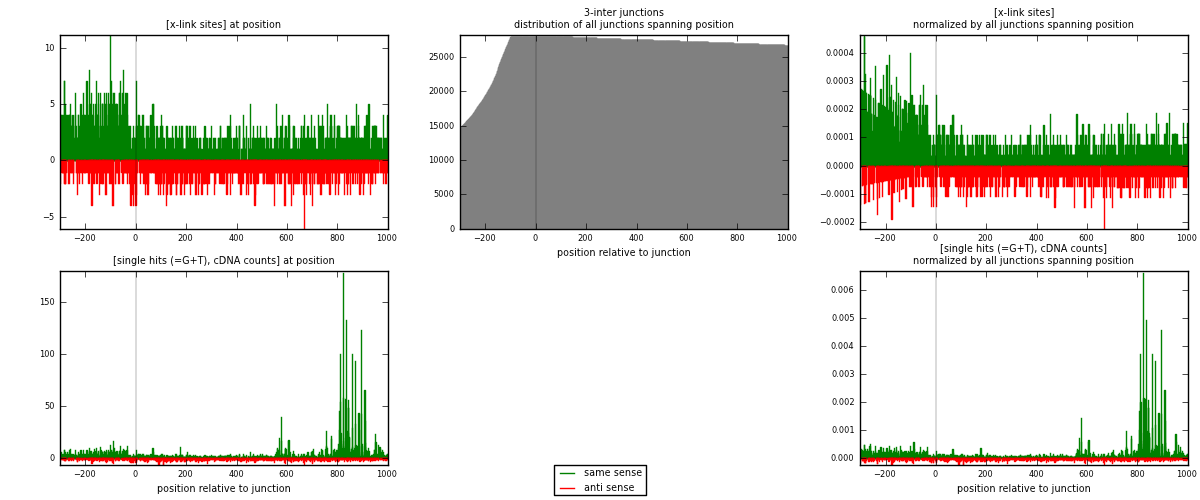
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2077 | 1.14% | 8.42% | 65.77% | 1090 | 1.18618 |
| anti | 1081 | 0.59% | 4.38% | 34.23% | 679 | 0.617362 |
| both | 3158 | 1.74% | 12.80% | 100.00% | 1751 | 1.80354 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4569 | 2.29% | 14.87% | 79.70% | 1090 | 2.60937 |
| anti | 1164 | 0.58% | 3.79% | 20.30% | 679 | 0.664763 |
| both | 5733 | 2.87% | 18.66% | 100.00% | 1751 | 3.27413 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
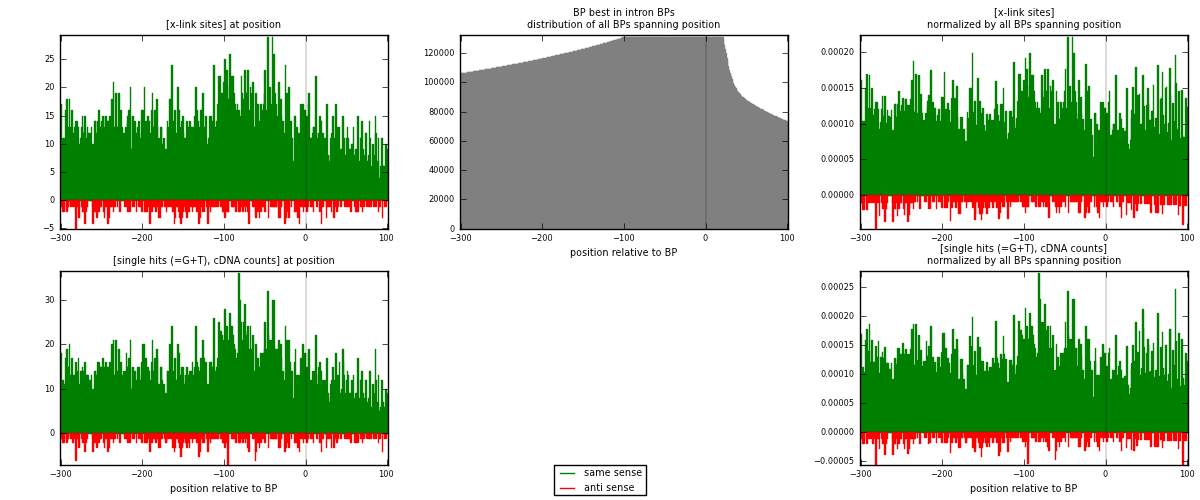
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5147 | 2.83% | 20.87% | 92.66% | 3576 | 1.31368 |
| anti | 408 | 0.22% | 1.65% | 7.34% | 352 | 0.104135 |
| both | 5555 | 3.06% | 22.52% | 100.00% | 3918 | 1.41782 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 5574 | 2.79% | 18.15% | 92.84% | 3576 | 1.42266 |
| anti | 430 | 0.22% | 1.40% | 7.16% | 352 | 0.10975 |
| both | 6004 | 3.01% | 19.55% | 100.00% | 3918 | 1.53241 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.