RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
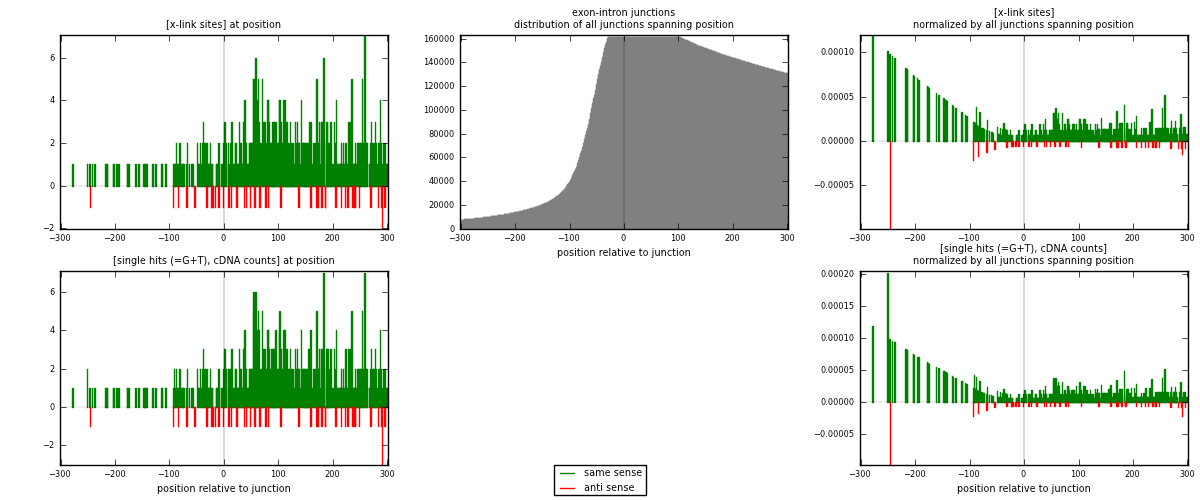 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 563 | 3.90% | 27.13% | 92.60% | 504 | 1.0255 |
| anti | 45 | 0.31% | 2.17% | 7.40% | 45 | 0.0819672 |
| both | 608 | 4.21% | 29.30% | 100.00% | 549 | 1.10747 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 578 | 3.84% | 24.88% | 92.63% | 504 | 1.05282 |
| anti | 46 | 0.31% | 1.98% | 7.37% | 45 | 0.0837887 |
| both | 624 | 4.15% | 26.86% | 100.00% | 549 | 1.13661 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
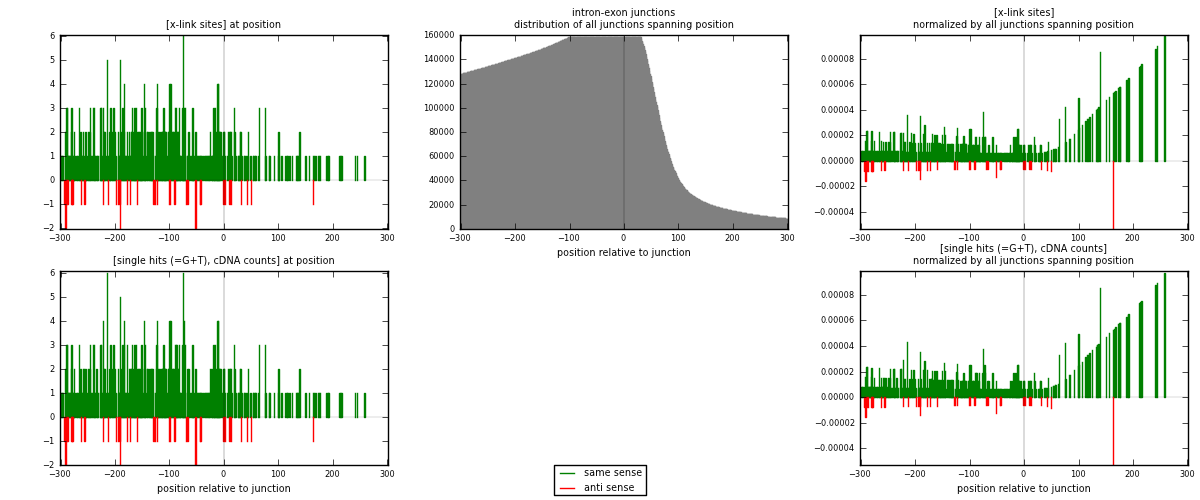 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 455 | 3.15% | 21.93% | 91.92% | 420 | 0.991285 |
| anti | 40 | 0.28% | 1.93% | 8.08% | 39 | 0.087146 |
| both | 495 | 3.43% | 23.86% | 100.00% | 459 | 1.07843 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 466 | 3.10% | 20.06% | 92.09% | 420 | 1.01525 |
| anti | 40 | 0.27% | 1.72% | 7.91% | 39 | 0.087146 |
| both | 506 | 3.36% | 21.78% | 100.00% | 459 | 1.1024 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
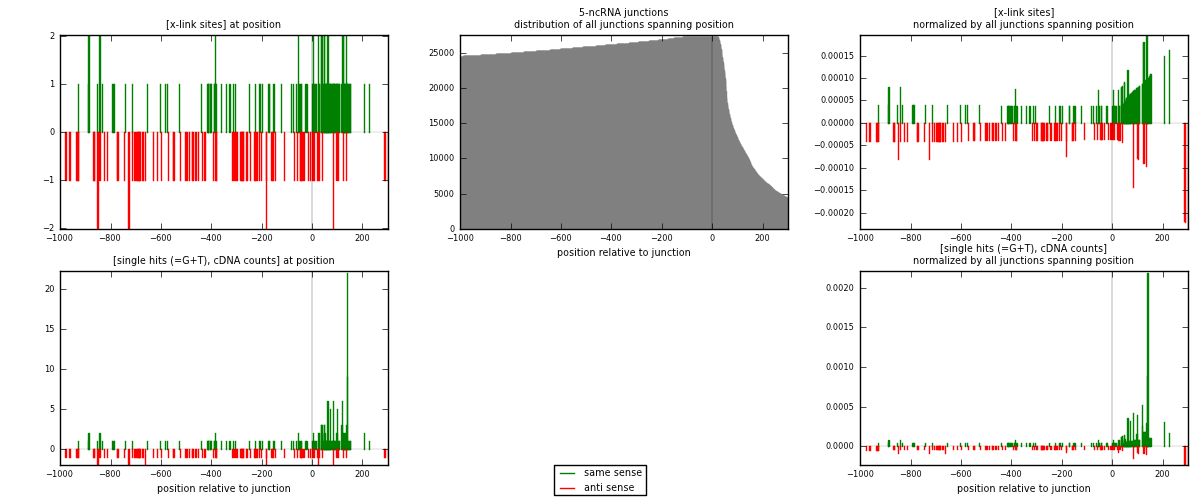 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 143 | 0.99% | 6.89% | 57.89% | 65 | 0.899371 |
| anti | 104 | 0.72% | 5.01% | 42.11% | 94 | 0.654088 |
| both | 247 | 1.71% | 11.90% | 100.00% | 159 | 1.55346 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 211 | 1.40% | 9.08% | 66.56% | 65 | 1.32704 |
| anti | 106 | 0.70% | 4.56% | 33.44% | 94 | 0.666667 |
| both | 317 | 2.11% | 13.65% | 100.00% | 159 | 1.99371 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
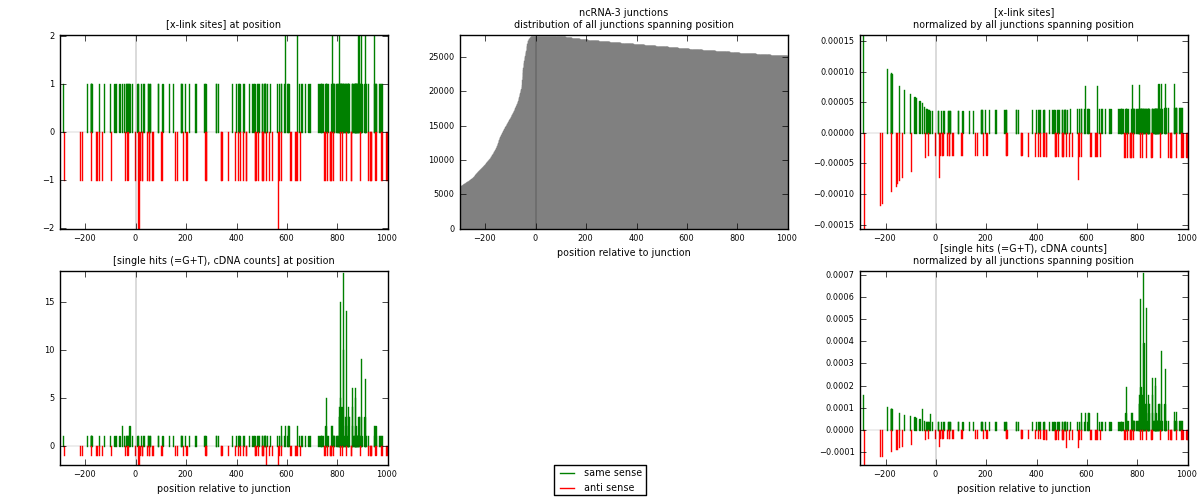 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 177 | 1.23% | 8.53% | 68.87% | 71 | 1.26429 |
| anti | 80 | 0.55% | 3.86% | 31.13% | 70 | 0.571429 |
| both | 257 | 1.78% | 12.39% | 100.00% | 140 | 1.83571 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 309 | 2.05% | 13.30% | 79.23% | 71 | 2.20714 |
| anti | 81 | 0.54% | 3.49% | 20.77% | 70 | 0.578571 |
| both | 390 | 2.59% | 16.79% | 100.00% | 140 | 2.78571 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
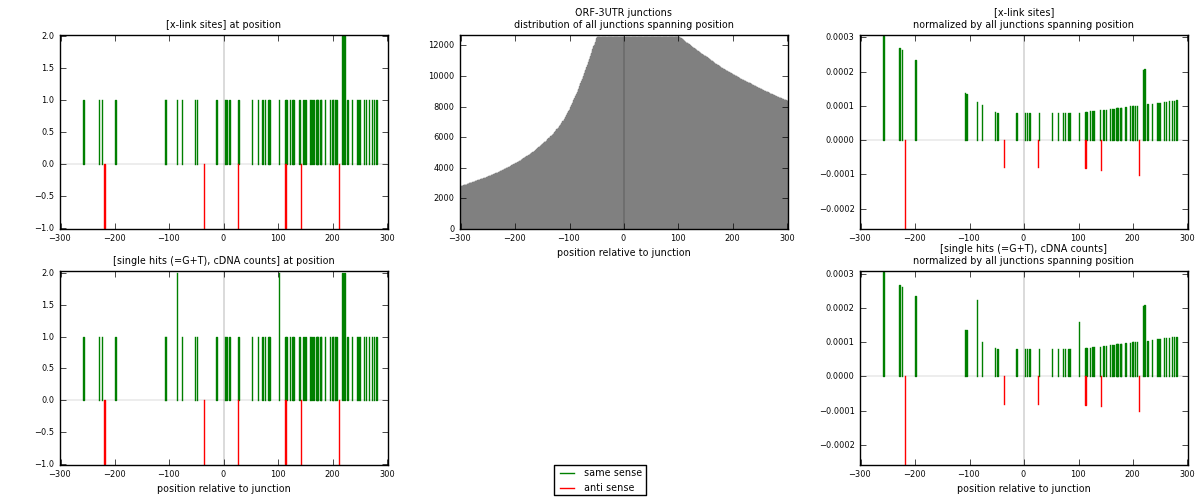
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 59 | 0.41% | 2.84% | 90.77% | 55 | 0.967213 |
| anti | 6 | 0.04% | 0.29% | 9.23% | 6 | 0.0983607 |
| both | 65 | 0.45% | 3.13% | 100.00% | 61 | 1.06557 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 61 | 0.41% | 2.63% | 91.04% | 55 | 1 |
| anti | 6 | 0.04% | 0.26% | 8.96% | 6 | 0.0983607 |
| both | 67 | 0.45% | 2.88% | 100.00% | 61 | 1.09836 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
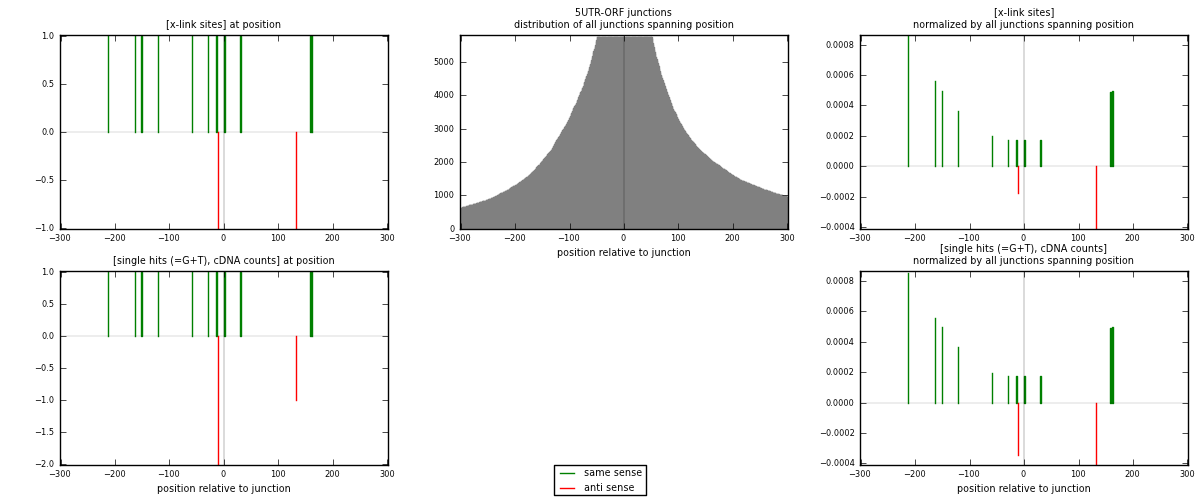
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 11 | 0.08% | 0.53% | 84.62% | 11 | 0.846154 |
| anti | 2 | 0.01% | 0.10% | 15.38% | 2 | 0.153846 |
| both | 13 | 0.09% | 0.63% | 100.00% | 13 | 1 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 11 | 0.07% | 0.47% | 78.57% | 11 | 0.846154 |
| anti | 3 | 0.02% | 0.13% | 21.43% | 2 | 0.230769 |
| both | 14 | 0.09% | 0.60% | 100.00% | 13 | 1.07692 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
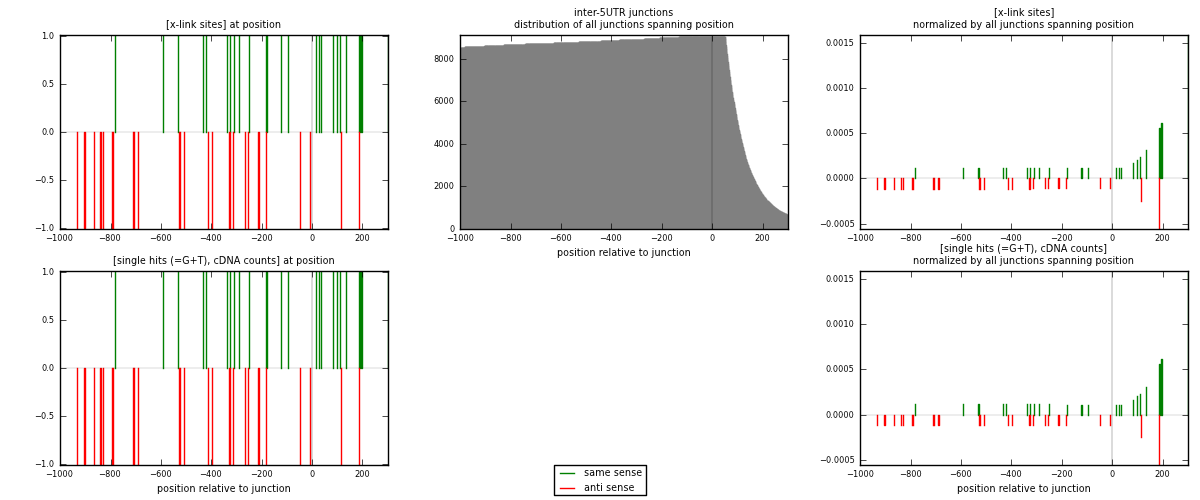
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 23 | 0.16% | 1.11% | 47.92% | 22 | 0.511111 |
| anti | 25 | 0.17% | 1.20% | 52.08% | 23 | 0.555556 |
| both | 48 | 0.33% | 2.31% | 100.00% | 45 | 1.06667 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 23 | 0.15% | 0.99% | 47.92% | 22 | 0.511111 |
| anti | 25 | 0.17% | 1.08% | 52.08% | 23 | 0.555556 |
| both | 48 | 0.32% | 2.07% | 100.00% | 45 | 1.06667 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
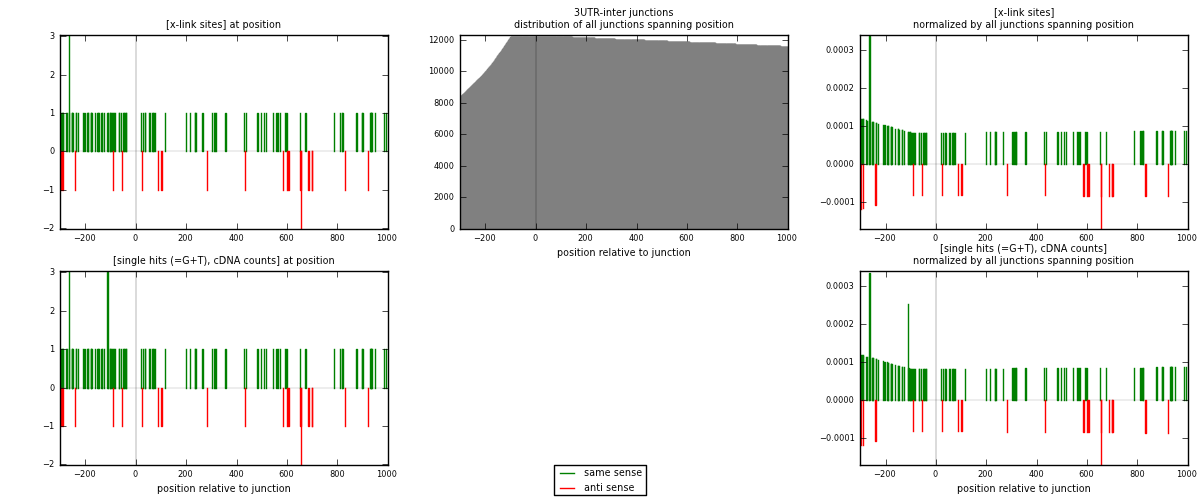
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 100 | 0.69% | 4.82% | 82.64% | 87 | 0.934579 |
| anti | 21 | 0.15% | 1.01% | 17.36% | 20 | 0.196262 |
| both | 121 | 0.84% | 5.83% | 100.00% | 107 | 1.13084 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 102 | 0.68% | 4.39% | 82.93% | 87 | 0.953271 |
| anti | 21 | 0.14% | 0.90% | 17.07% | 20 | 0.196262 |
| both | 123 | 0.82% | 5.29% | 100.00% | 107 | 1.14953 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
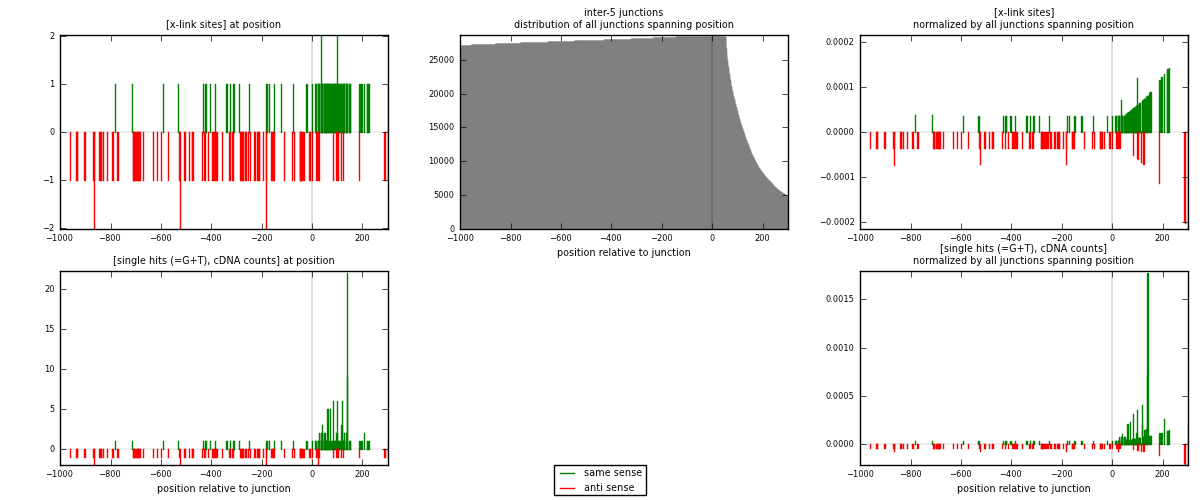
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 94 | 0.65% | 4.53% | 47.96% | 45 | 0.696296 |
| anti | 102 | 0.71% | 4.92% | 52.04% | 90 | 0.755556 |
| both | 196 | 1.36% | 9.45% | 100.00% | 135 | 1.45185 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 158 | 1.05% | 6.80% | 60.54% | 45 | 1.17037 |
| anti | 103 | 0.68% | 4.43% | 39.46% | 90 | 0.762963 |
| both | 261 | 1.73% | 11.24% | 100.00% | 135 | 1.93333 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
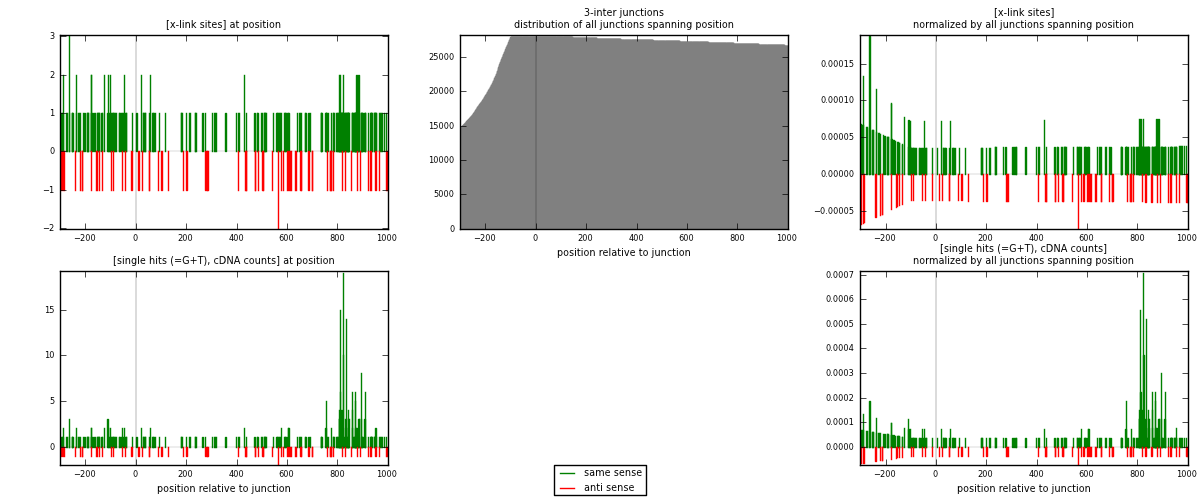
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 232 | 1.61% | 11.18% | 76.57% | 115 | 1.31073 |
| anti | 71 | 0.49% | 3.42% | 23.43% | 62 | 0.40113 |
| both | 303 | 2.10% | 14.60% | 100.00% | 177 | 1.71186 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 365 | 2.43% | 15.71% | 83.72% | 115 | 2.06215 |
| anti | 71 | 0.47% | 3.06% | 16.28% | 62 | 0.40113 |
| both | 436 | 2.90% | 18.77% | 100.00% | 177 | 2.46328 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
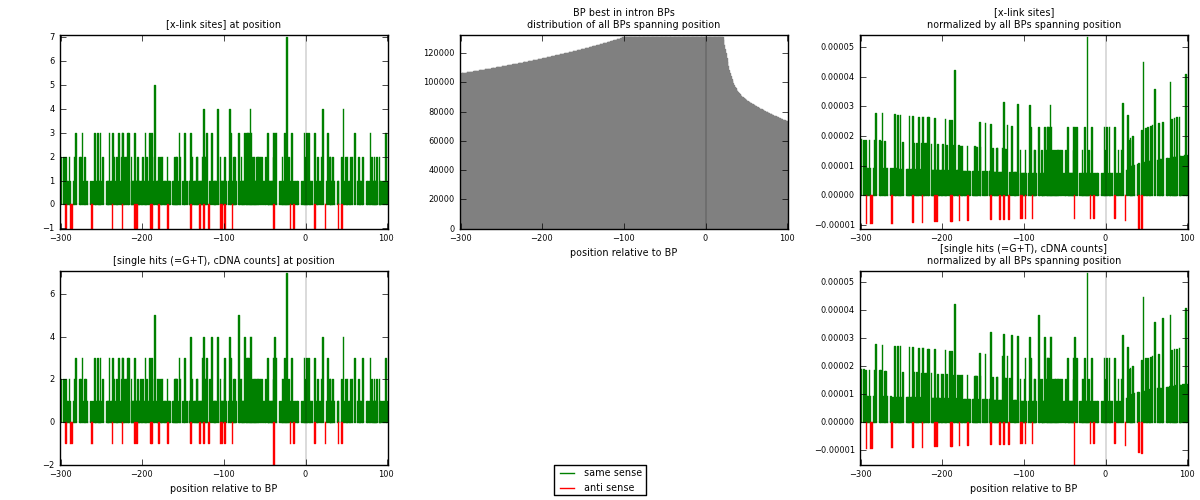
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 442 | 3.06% | 21.30% | 94.04% | 400 | 1.03513 |
| anti | 28 | 0.19% | 1.35% | 5.96% | 27 | 0.0655738 |
| both | 470 | 3.26% | 22.65% | 100.00% | 427 | 1.1007 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 461 | 3.06% | 19.85% | 94.08% | 400 | 1.07963 |
| anti | 29 | 0.19% | 1.25% | 5.92% | 27 | 0.0679157 |
| both | 490 | 3.26% | 21.09% | 100.00% | 427 | 1.14754 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.