RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
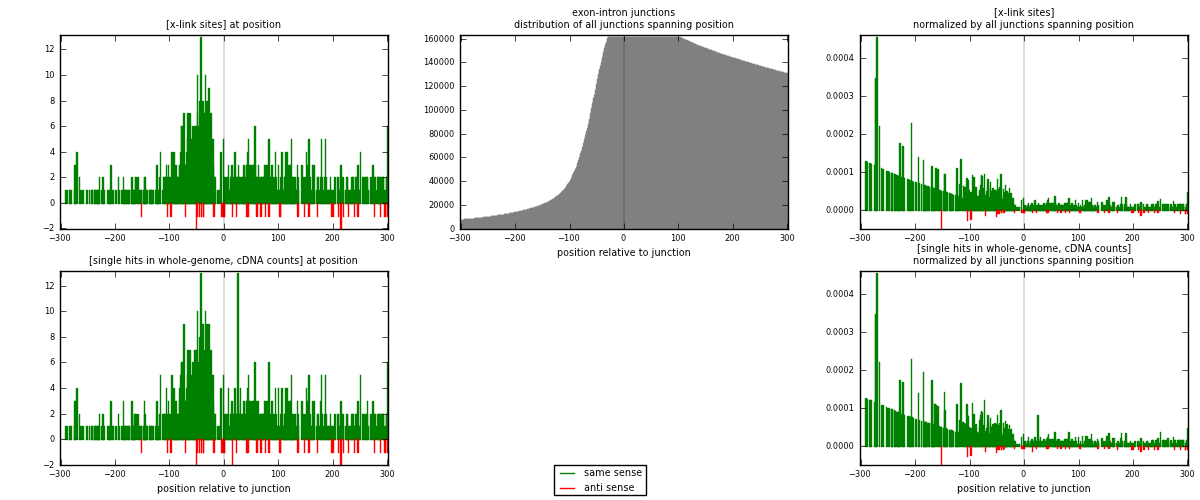 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 946 | 4.76% | 16.22% | 95.46% | 831 | 1.08486 |
| anti | 45 | 0.23% | 0.77% | 4.54% | 41 | 0.0516055 |
| both | 991 | 4.98% | 16.99% | 100.00% | 872 | 1.13647 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 986 | 4.32% | 13.79% | 95.54% | 831 | 1.13073 |
| anti | 46 | 0.20% | 0.64% | 4.46% | 41 | 0.0527523 |
| both | 1032 | 4.53% | 14.44% | 100.00% | 872 | 1.18349 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
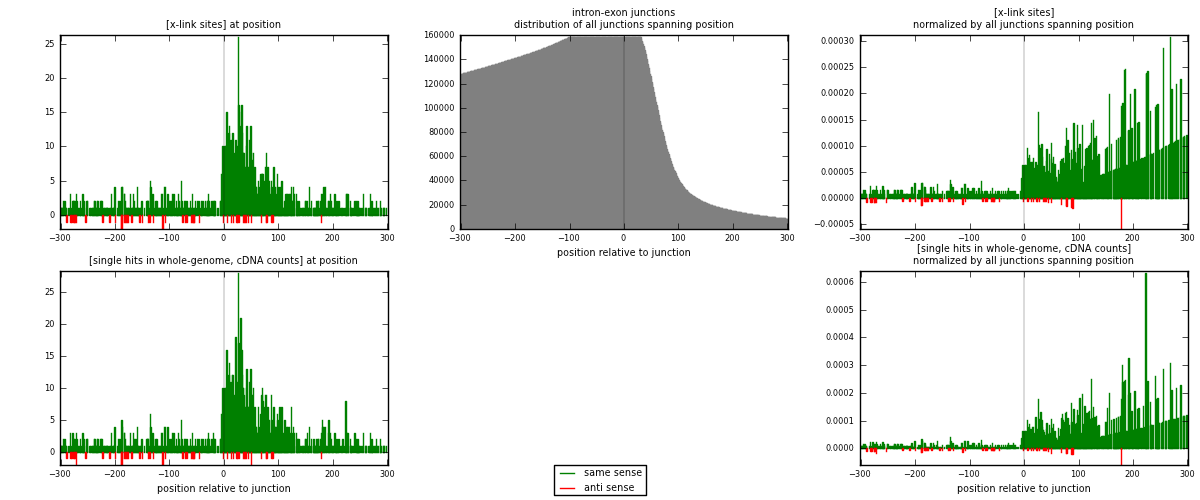 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1221 | 6.14% | 20.93% | 95.47% | 1025 | 1.1337 |
| anti | 58 | 0.29% | 0.99% | 4.53% | 52 | 0.0538533 |
| both | 1279 | 6.43% | 21.92% | 100.00% | 1077 | 1.18756 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1330 | 5.83% | 18.60% | 95.68% | 1025 | 1.23491 |
| anti | 60 | 0.26% | 0.84% | 4.32% | 52 | 0.0557103 |
| both | 1390 | 6.10% | 19.44% | 100.00% | 1077 | 1.29062 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
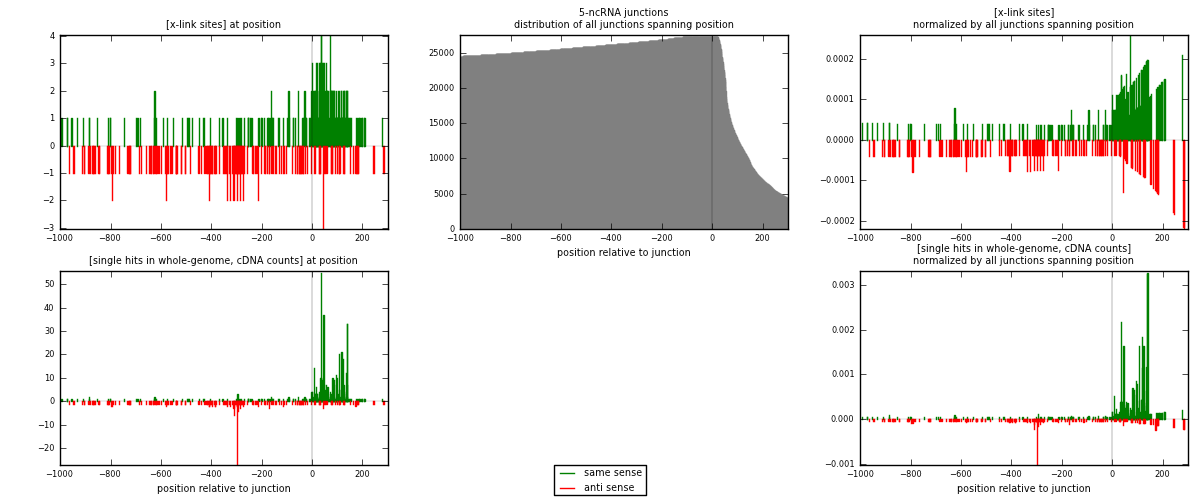 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 264 | 1.33% | 4.53% | 58.54% | 103 | 1.26316 |
| anti | 187 | 0.94% | 3.21% | 41.46% | 108 | 0.894737 |
| both | 451 | 2.27% | 7.73% | 100.00% | 209 | 2.15789 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 661 | 2.90% | 9.25% | 73.94% | 103 | 3.16268 |
| anti | 233 | 1.02% | 3.26% | 26.06% | 108 | 1.11483 |
| both | 894 | 3.92% | 12.51% | 100.00% | 209 | 4.27751 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
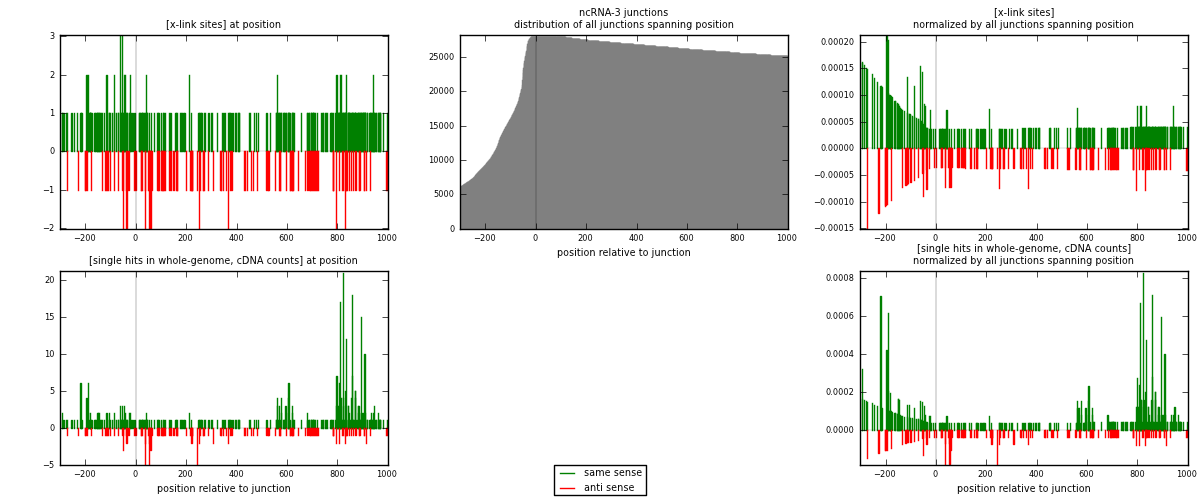 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 285 | 1.43% | 4.89% | 67.70% | 112 | 1.31944 |
| anti | 136 | 0.68% | 2.33% | 32.30% | 104 | 0.62963 |
| both | 421 | 2.12% | 7.22% | 100.00% | 216 | 1.94907 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 488 | 2.14% | 6.83% | 76.01% | 112 | 2.25926 |
| anti | 154 | 0.68% | 2.15% | 23.99% | 104 | 0.712963 |
| both | 642 | 2.82% | 8.98% | 100.00% | 216 | 2.97222 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
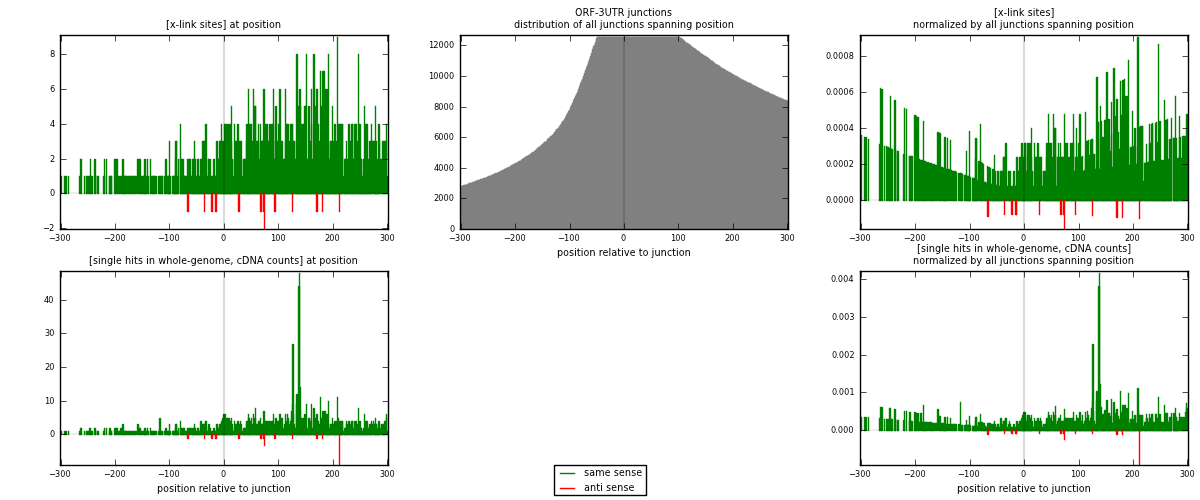 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1058 | 5.32% | 18.14% | 98.60% | 563 | 1.84642 |
| anti | 15 | 0.08% | 0.26% | 1.40% | 10 | 0.026178 |
| both | 1073 | 5.39% | 18.39% | 100.00% | 573 | 1.8726 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1304 | 5.72% | 18.24% | 98.19% | 563 | 2.27574 |
| anti | 24 | 0.11% | 0.34% | 1.81% | 10 | 0.0418848 |
| both | 1328 | 5.82% | 18.58% | 100.00% | 573 | 2.31763 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
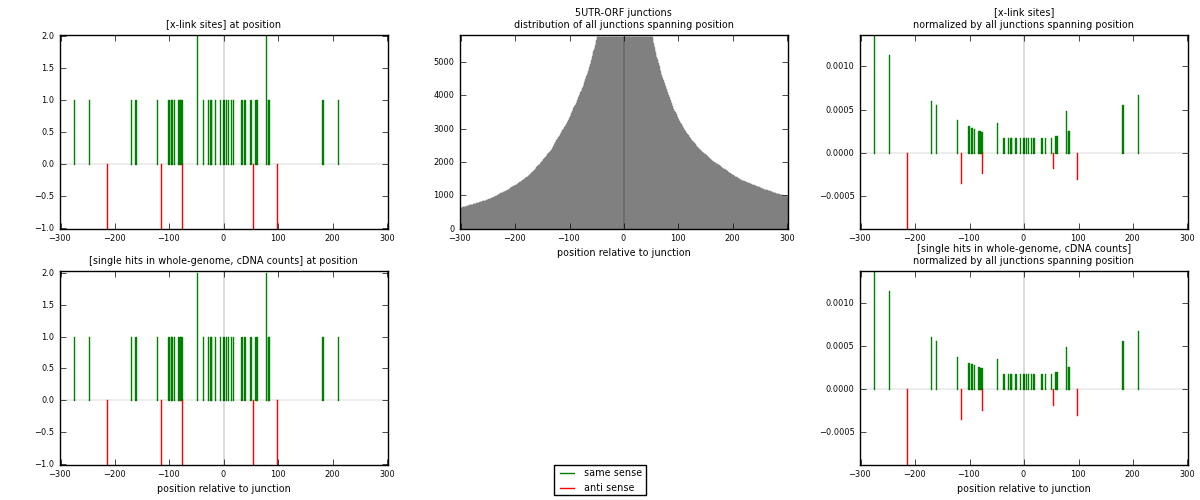 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 37 | 0.19% | 0.63% | 88.10% | 30 | 1.05714 |
| anti | 5 | 0.03% | 0.09% | 11.90% | 5 | 0.142857 |
| both | 42 | 0.21% | 0.72% | 100.00% | 35 | 1.2 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 37 | 0.16% | 0.52% | 88.10% | 30 | 1.05714 |
| anti | 5 | 0.02% | 0.07% | 11.90% | 5 | 0.142857 |
| both | 42 | 0.18% | 0.59% | 100.00% | 35 | 1.2 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
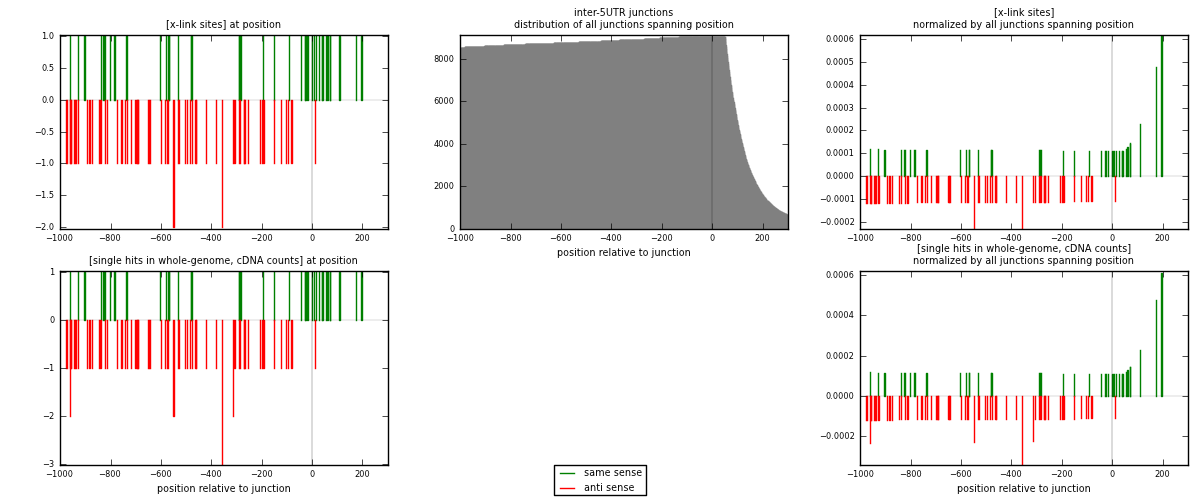 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 40 | 0.20% | 0.69% | 38.83% | 29 | 0.481928 |
| anti | 63 | 0.32% | 1.08% | 61.17% | 55 | 0.759036 |
| both | 103 | 0.52% | 1.77% | 100.00% | 83 | 1.24096 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 40 | 0.18% | 0.56% | 37.74% | 29 | 0.481928 |
| anti | 66 | 0.29% | 0.92% | 62.26% | 55 | 0.795181 |
| both | 106 | 0.46% | 1.48% | 100.00% | 83 | 1.27711 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
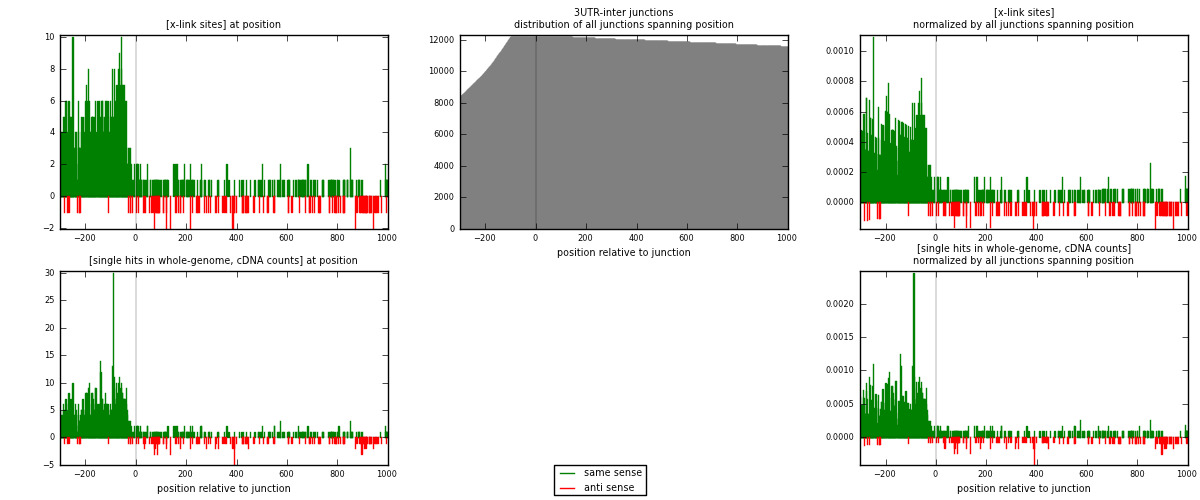 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1160 | 5.83% | 19.88% | 89.37% | 633 | 1.65714 |
| anti | 138 | 0.69% | 2.37% | 10.63% | 69 | 0.197143 |
| both | 1298 | 6.53% | 22.25% | 100.00% | 700 | 1.85429 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1374 | 6.03% | 19.22% | 89.69% | 633 | 1.96286 |
| anti | 158 | 0.69% | 2.21% | 10.31% | 69 | 0.225714 |
| both | 1532 | 6.72% | 21.43% | 100.00% | 700 | 2.18857 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
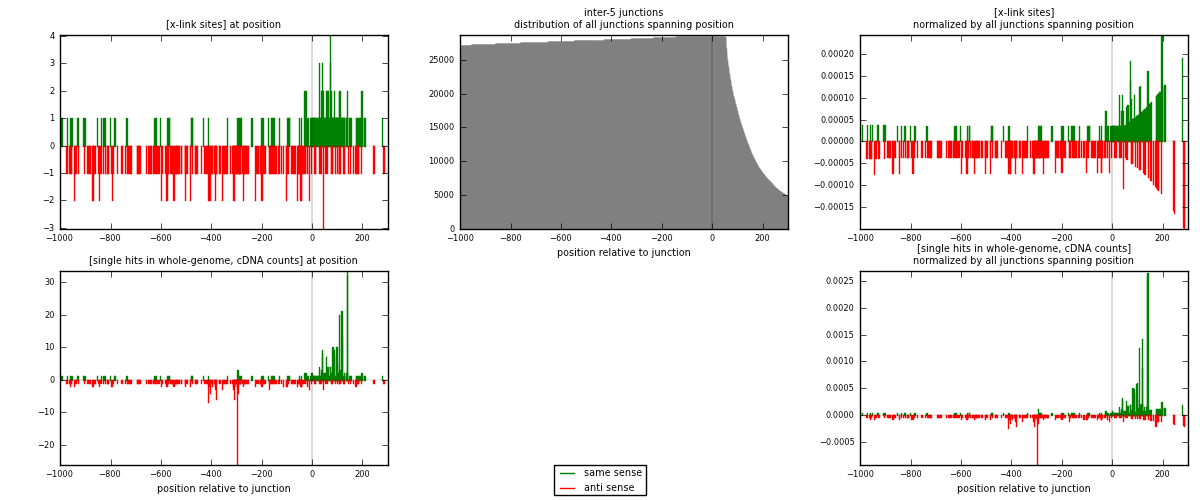 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 165 | 0.83% | 2.83% | 38.64% | 72 | 0.743243 |
| anti | 262 | 1.32% | 4.49% | 61.36% | 151 | 1.18018 |
| both | 427 | 2.15% | 7.32% | 100.00% | 222 | 1.92342 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 362 | 1.59% | 5.06% | 52.54% | 72 | 1.63063 |
| anti | 327 | 1.43% | 4.57% | 47.46% | 151 | 1.47297 |
| both | 689 | 3.02% | 9.64% | 100.00% | 222 | 3.1036 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
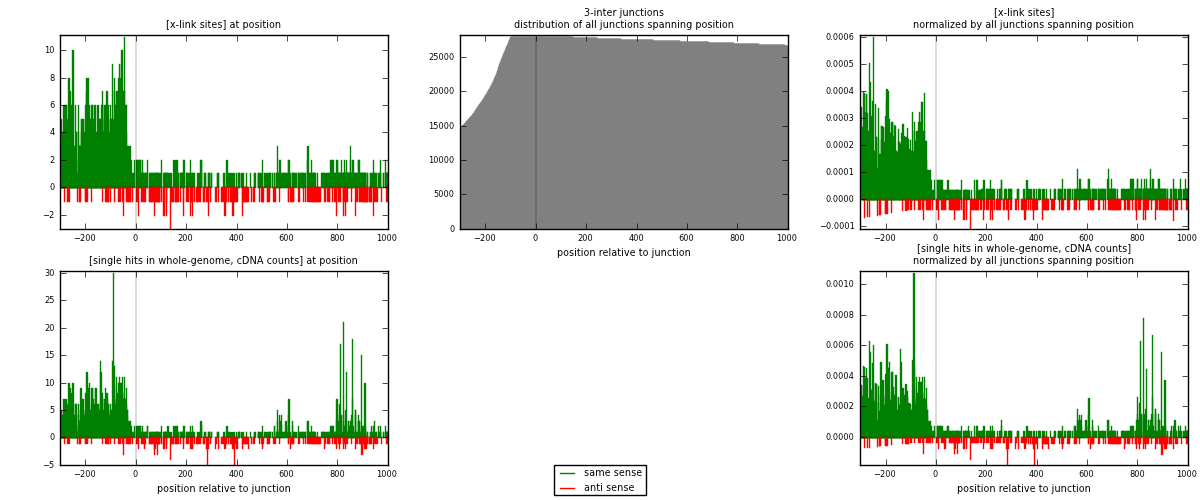 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1447 | 7.28% | 24.80% | 85.37% | 732 | 1.66513 |
| anti | 248 | 1.25% | 4.25% | 14.63% | 140 | 0.285386 |
| both | 1695 | 8.52% | 29.05% | 100.00% | 869 | 1.95052 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1865 | 8.18% | 26.09% | 87.03% | 732 | 2.14614 |
| anti | 278 | 1.22% | 3.89% | 12.97% | 140 | 0.319908 |
| both | 2143 | 9.40% | 29.98% | 100.00% | 869 | 2.46605 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
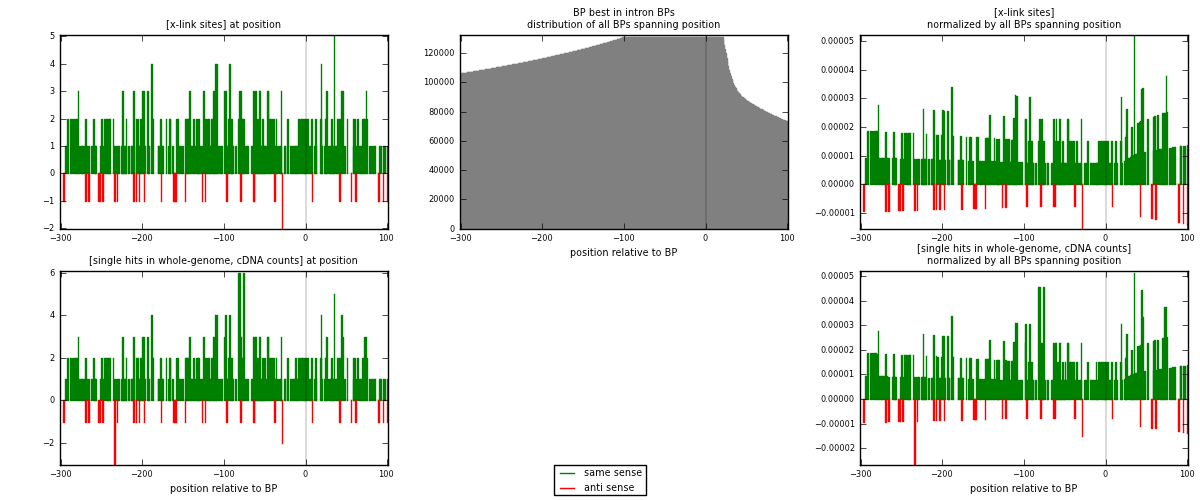 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 390 | 1.96% | 6.68% | 91.76% | 325 | 1.09859 |
| anti | 35 | 0.18% | 0.60% | 8.24% | 31 | 0.0985915 |
| both | 425 | 2.14% | 7.28% | 100.00% | 355 | 1.19718 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 408 | 1.79% | 5.71% | 91.69% | 325 | 1.1493 |
| anti | 37 | 0.16% | 0.52% | 8.31% | 31 | 0.104225 |
| both | 445 | 1.95% | 6.22% | 100.00% | 355 | 1.25352 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.