RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
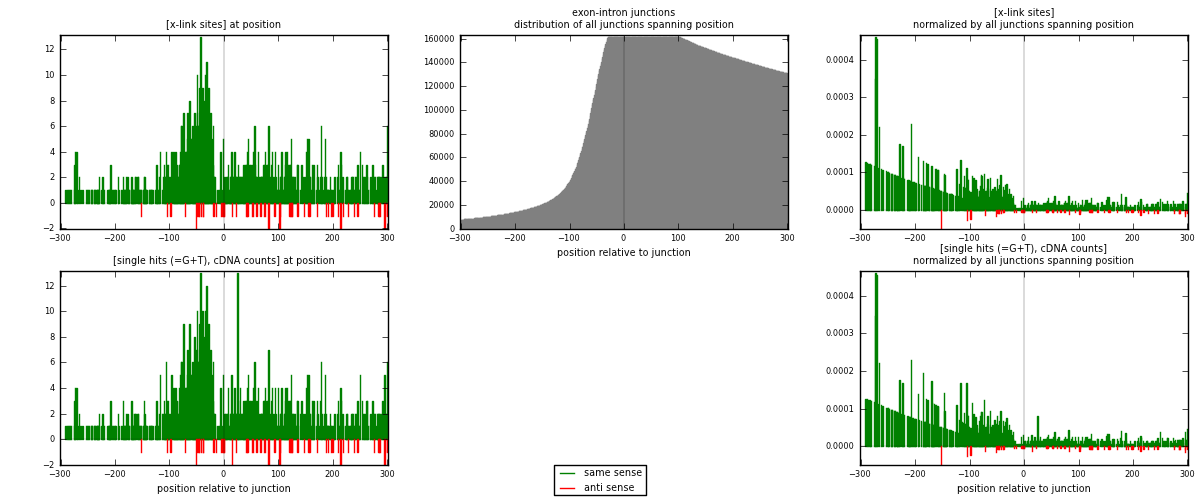 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1017 | 4.57% | 15.89% | 94.17% | 891 | 1.07278 |
| anti | 63 | 0.28% | 0.98% | 5.83% | 57 | 0.0664557 |
| both | 1080 | 4.85% | 16.88% | 100.00% | 948 | 1.13924 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1065 | 4.10% | 13.55% | 94.33% | 891 | 1.12342 |
| anti | 64 | 0.25% | 0.81% | 5.67% | 57 | 0.0675105 |
| both | 1129 | 4.35% | 14.36% | 100.00% | 948 | 1.19093 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
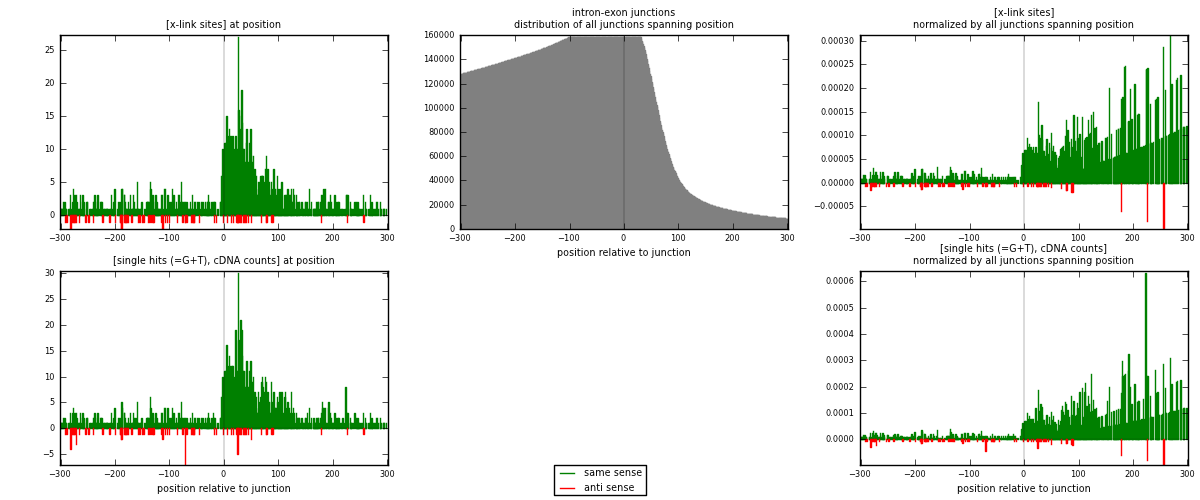 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1286 | 5.78% | 20.09% | 93.87% | 1077 | 1.11535 |
| anti | 84 | 0.38% | 1.31% | 6.13% | 76 | 0.0728534 |
| both | 1370 | 6.16% | 21.41% | 100.00% | 1153 | 1.1882 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1404 | 5.41% | 17.86% | 93.41% | 1077 | 1.21769 |
| anti | 99 | 0.38% | 1.26% | 6.59% | 76 | 0.085863 |
| both | 1503 | 5.79% | 19.12% | 100.00% | 1153 | 1.30356 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
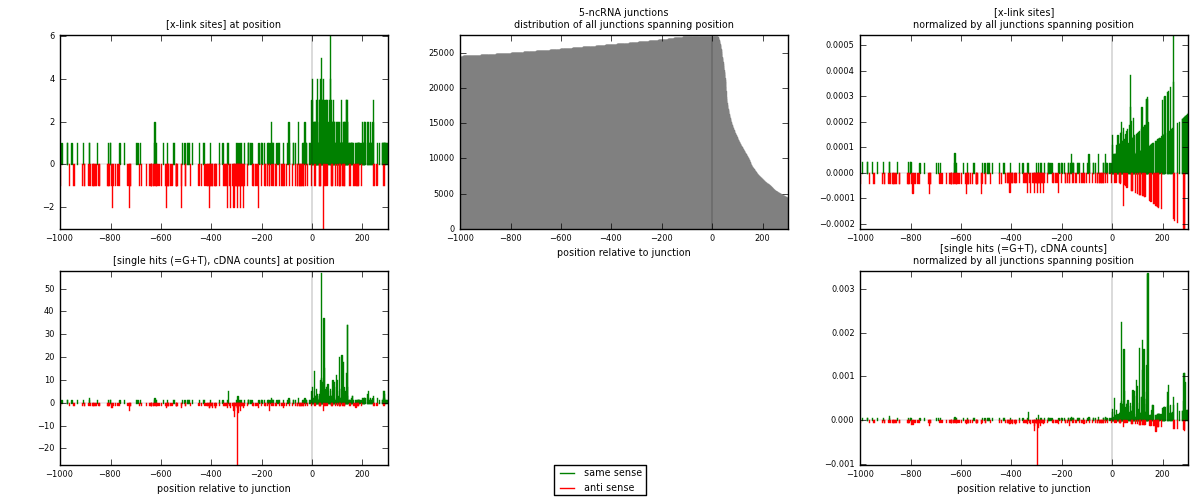 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 357 | 1.60% | 5.58% | 63.64% | 118 | 1.51271 |
| anti | 204 | 0.92% | 3.19% | 36.36% | 121 | 0.864407 |
| both | 561 | 2.52% | 8.77% | 100.00% | 236 | 2.37712 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 813 | 3.13% | 10.34% | 76.41% | 118 | 3.44492 |
| anti | 251 | 0.97% | 3.19% | 23.59% | 121 | 1.06356 |
| both | 1064 | 4.10% | 13.53% | 100.00% | 236 | 4.50847 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
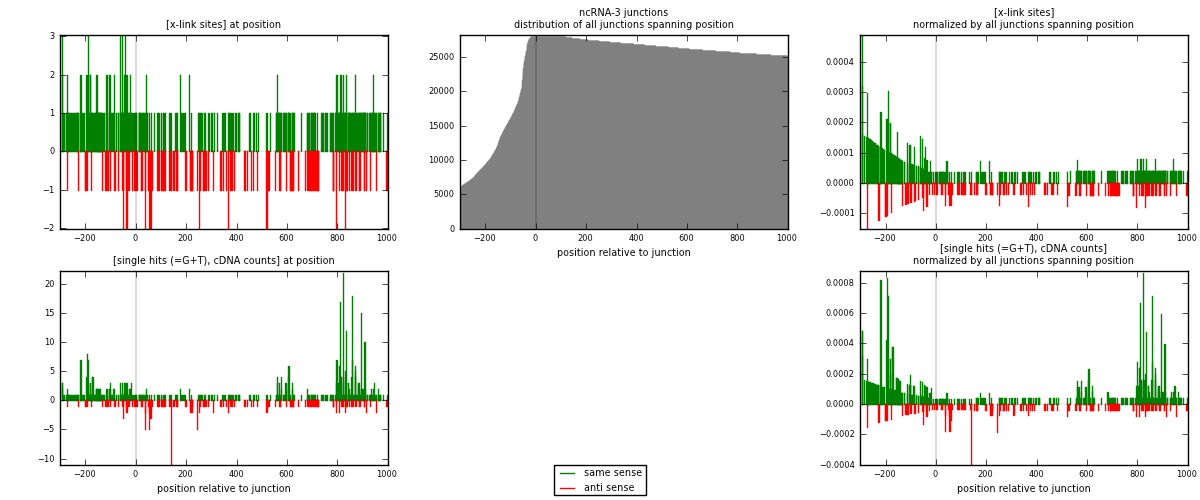 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 350 | 1.57% | 5.47% | 70.14% | 127 | 1.45228 |
| anti | 149 | 0.67% | 2.33% | 29.86% | 115 | 0.618257 |
| both | 499 | 2.24% | 7.80% | 100.00% | 241 | 2.07054 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 576 | 2.22% | 7.33% | 76.29% | 127 | 2.39004 |
| anti | 179 | 0.69% | 2.28% | 23.71% | 115 | 0.742739 |
| both | 755 | 2.91% | 9.60% | 100.00% | 241 | 3.13278 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
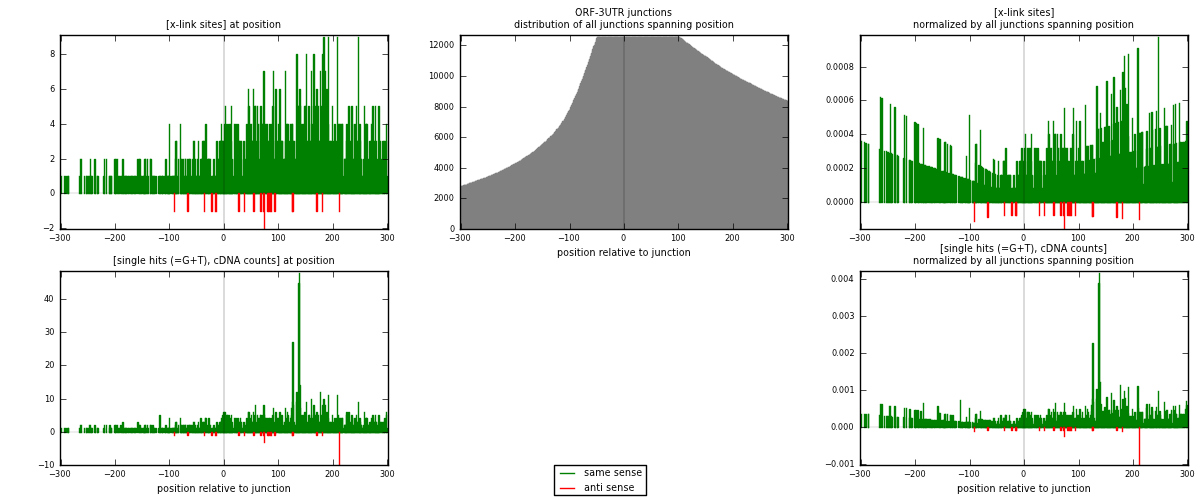 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1132 | 5.09% | 17.69% | 98.01% | 600 | 1.8527 |
| anti | 23 | 0.10% | 0.36% | 1.99% | 12 | 0.0376432 |
| both | 1155 | 5.19% | 18.05% | 100.00% | 611 | 1.89034 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1392 | 5.36% | 17.71% | 97.68% | 600 | 2.27823 |
| anti | 33 | 0.13% | 0.42% | 2.32% | 12 | 0.0540098 |
| both | 1425 | 5.49% | 18.13% | 100.00% | 611 | 2.33224 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
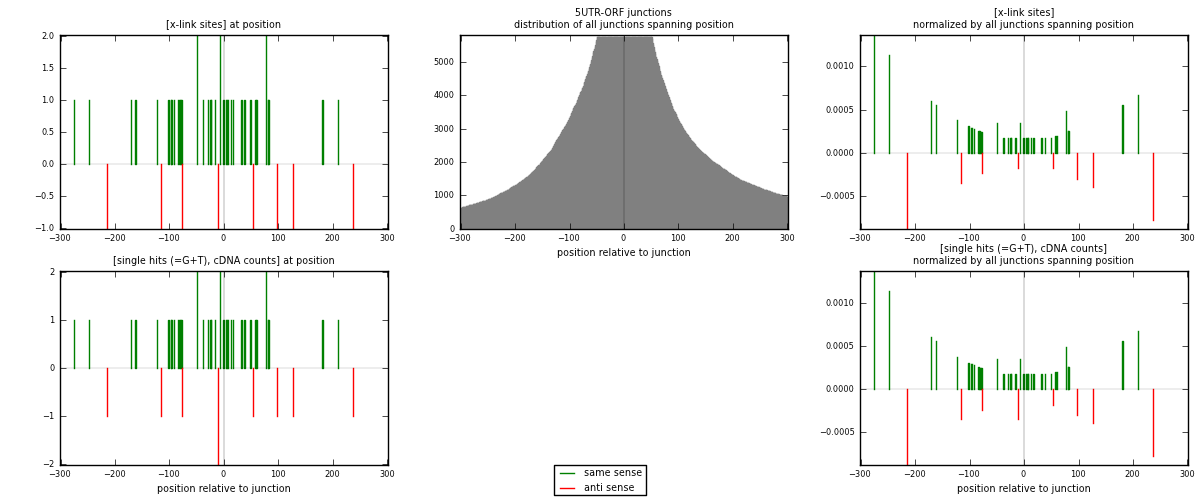 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 39 | 0.18% | 0.61% | 82.98% | 32 | 0.975 |
| anti | 8 | 0.04% | 0.12% | 17.02% | 8 | 0.2 |
| both | 47 | 0.21% | 0.73% | 100.00% | 40 | 1.175 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 39 | 0.15% | 0.50% | 81.25% | 32 | 0.975 |
| anti | 9 | 0.03% | 0.11% | 18.75% | 8 | 0.225 |
| both | 48 | 0.18% | 0.61% | 100.00% | 40 | 1.2 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
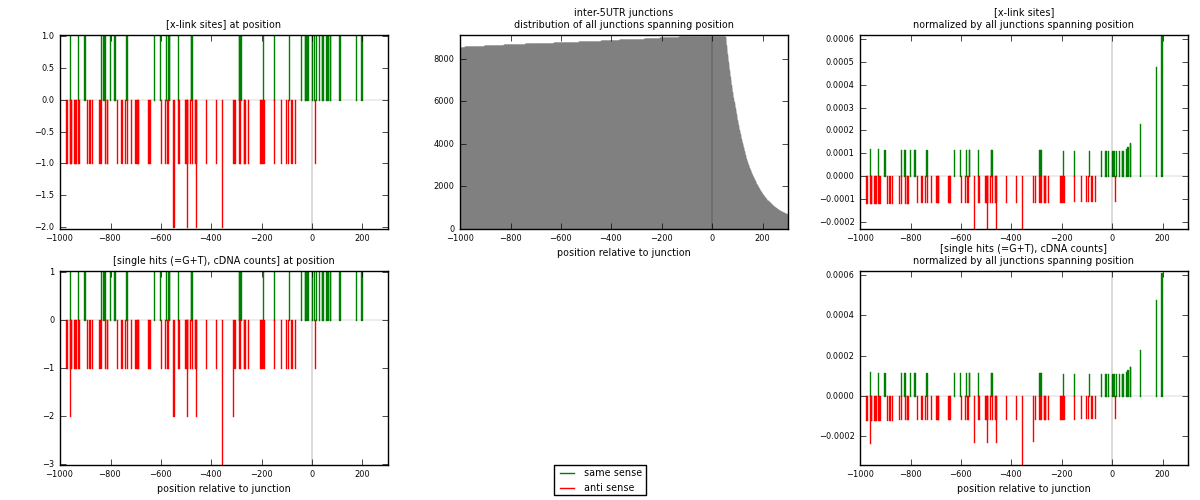 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 41 | 0.18% | 0.64% | 35.96% | 30 | 0.455556 |
| anti | 73 | 0.33% | 1.14% | 64.04% | 61 | 0.811111 |
| both | 114 | 0.51% | 1.78% | 100.00% | 90 | 1.26667 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 41 | 0.16% | 0.52% | 35.04% | 30 | 0.455556 |
| anti | 76 | 0.29% | 0.97% | 64.96% | 61 | 0.844444 |
| both | 117 | 0.45% | 1.49% | 100.00% | 90 | 1.3 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
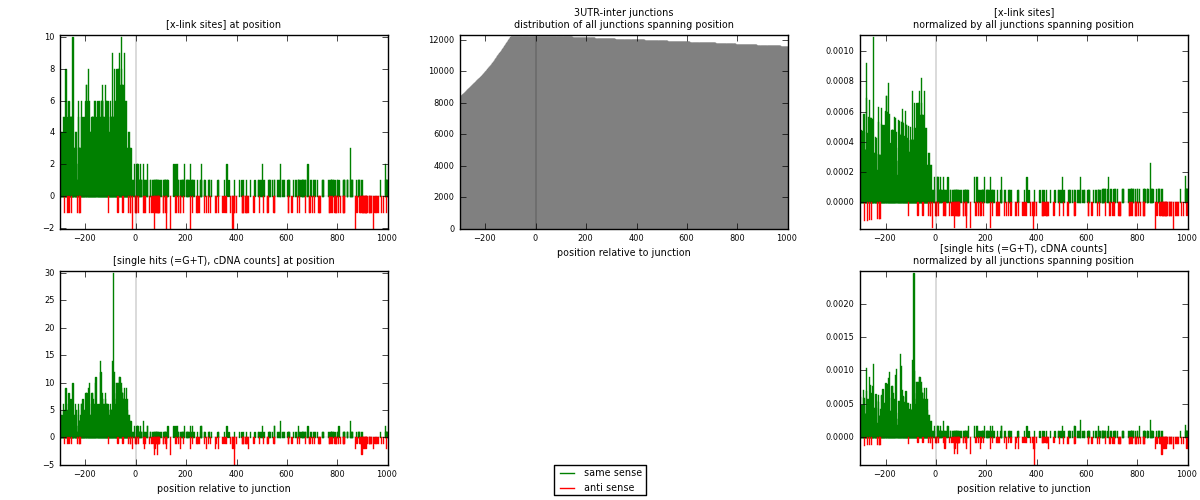 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1225 | 5.51% | 19.14% | 88.96% | 664 | 1.66214 |
| anti | 152 | 0.68% | 2.38% | 11.04% | 75 | 0.206242 |
| both | 1377 | 6.19% | 21.52% | 100.00% | 737 | 1.86839 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1447 | 5.57% | 18.40% | 89.27% | 664 | 1.96336 |
| anti | 174 | 0.67% | 2.21% | 10.73% | 75 | 0.236092 |
| both | 1621 | 6.24% | 20.62% | 100.00% | 737 | 2.19946 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
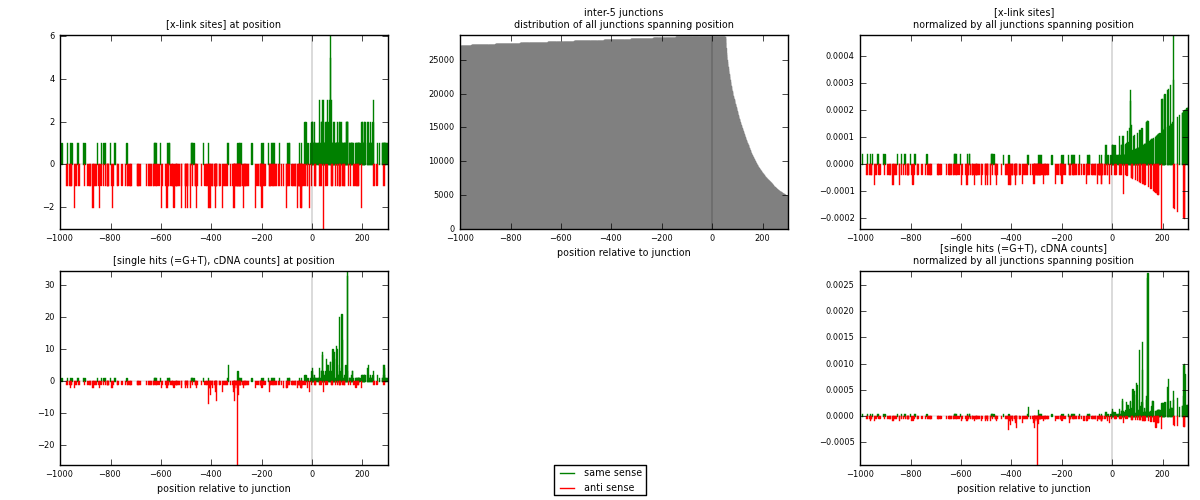
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 250 | 1.12% | 3.91% | 47.08% | 81 | 1.03306 |
| anti | 281 | 1.26% | 4.39% | 52.92% | 162 | 1.16116 |
| both | 531 | 2.39% | 8.30% | 100.00% | 242 | 2.19421 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 493 | 1.90% | 6.27% | 58.76% | 81 | 2.03719 |
| anti | 346 | 1.33% | 4.40% | 41.24% | 162 | 1.42975 |
| both | 839 | 3.23% | 10.67% | 100.00% | 242 | 3.46694 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
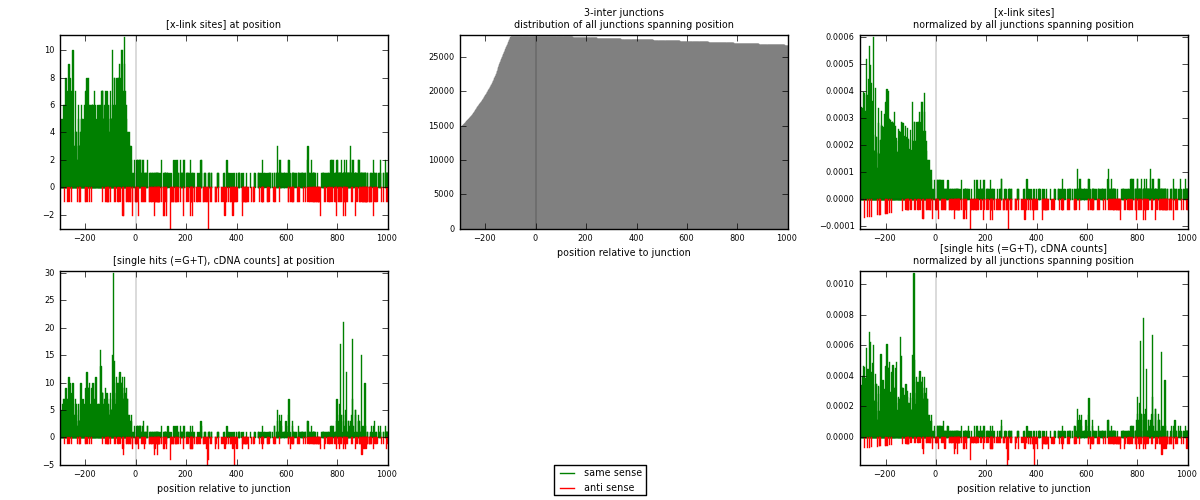
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1571 | 7.06% | 24.55% | 85.10% | 770 | 1.70575 |
| anti | 275 | 1.24% | 4.30% | 14.90% | 154 | 0.298588 |
| both | 1846 | 8.30% | 28.84% | 100.00% | 921 | 2.00434 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2019 | 7.78% | 25.68% | 86.73% | 770 | 2.19218 |
| anti | 309 | 1.19% | 3.93% | 13.27% | 154 | 0.335505 |
| both | 2328 | 8.97% | 29.61% | 100.00% | 921 | 2.52769 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
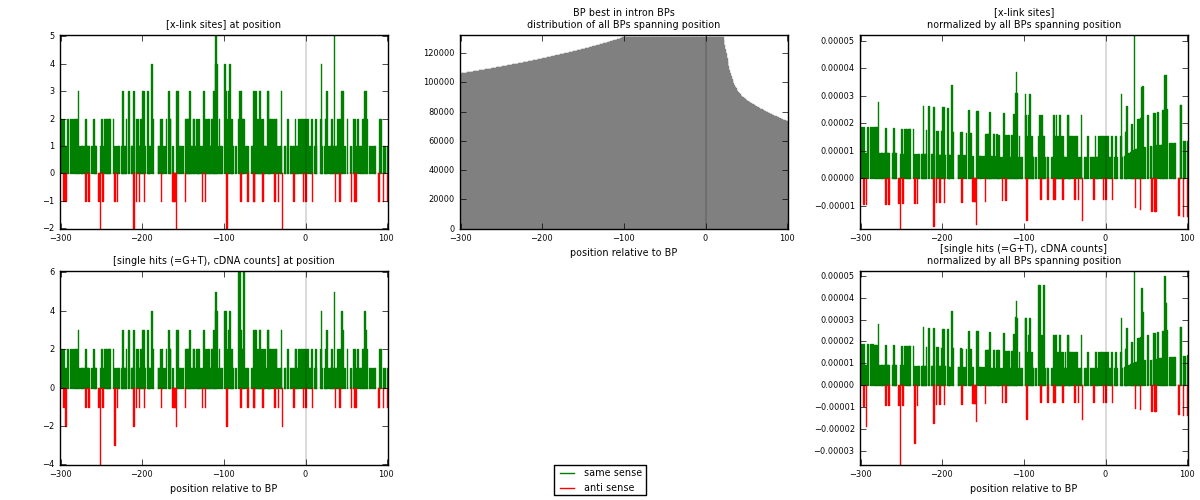
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 423 | 1.90% | 6.61% | 89.62% | 355 | 1.06281 |
| anti | 49 | 0.22% | 0.77% | 10.38% | 44 | 0.123116 |
| both | 472 | 2.12% | 7.38% | 100.00% | 398 | 1.18593 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 441 | 1.70% | 5.61% | 89.09% | 355 | 1.10804 |
| anti | 54 | 0.21% | 0.69% | 10.91% | 44 | 0.135678 |
| both | 495 | 1.91% | 6.30% | 100.00% | 398 | 1.24372 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.