RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
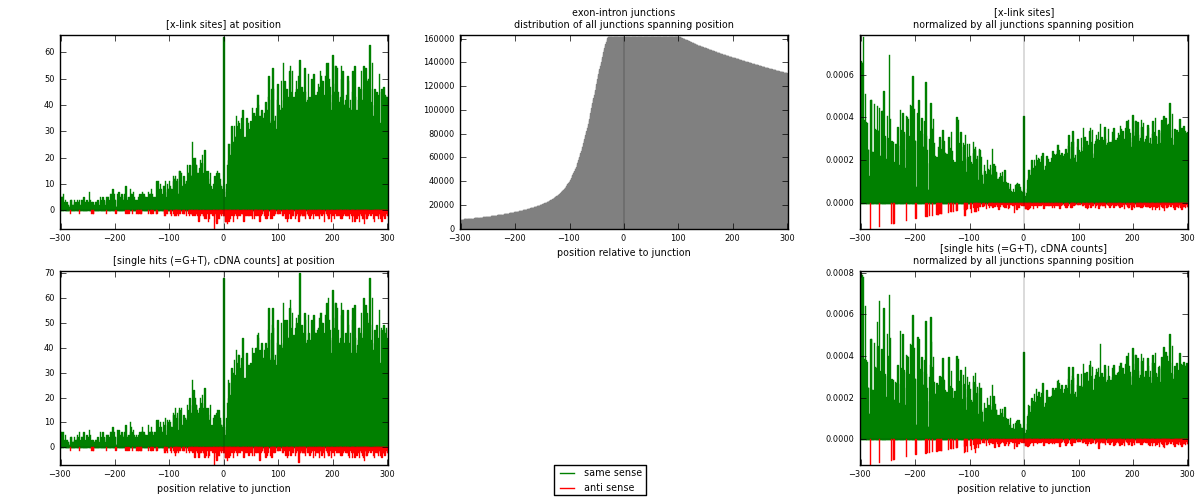 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13638 | 4.11% | 27.04% | 96.08% | 10097 | 1.29184 |
| anti | 557 | 0.17% | 1.10% | 3.92% | 486 | 0.0527612 |
| both | 14195 | 4.28% | 28.15% | 100.00% | 10557 | 1.34461 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 14297 | 4.05% | 25.98% | 96.09% | 10097 | 1.35427 |
| anti | 581 | 0.16% | 1.06% | 3.91% | 486 | 0.0550346 |
| both | 14878 | 4.22% | 27.03% | 100.00% | 10557 | 1.4093 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
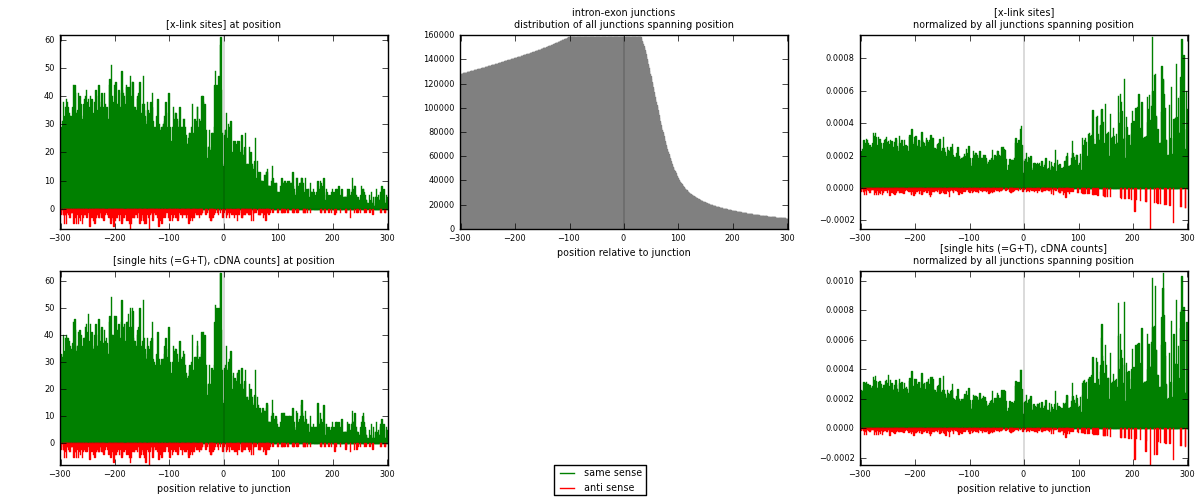 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12239 | 3.69% | 24.27% | 94.45% | 9320 | 1.23701 |
| anti | 719 | 0.22% | 1.43% | 5.55% | 606 | 0.0726703 |
| both | 12958 | 3.90% | 25.69% | 100.00% | 9894 | 1.30968 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 12875 | 3.65% | 23.39% | 94.53% | 9320 | 1.30129 |
| anti | 745 | 0.21% | 1.35% | 5.47% | 606 | 0.0752982 |
| both | 13620 | 3.86% | 24.75% | 100.00% | 9894 | 1.37659 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
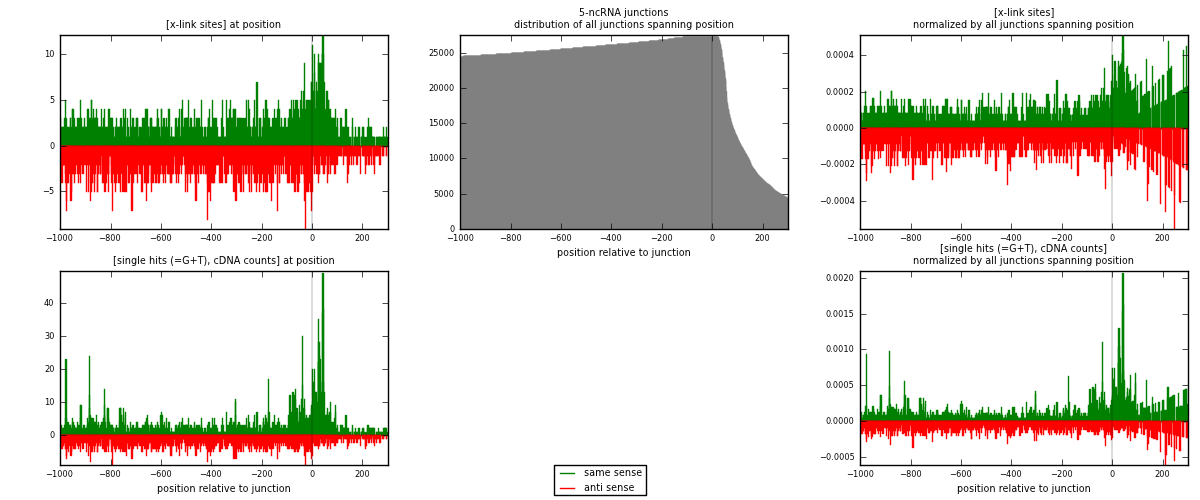 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2080 | 0.63% | 4.12% | 48.73% | 963 | 0.936094 |
| anti | 2188 | 0.66% | 4.34% | 51.27% | 1290 | 0.984698 |
| both | 4268 | 1.29% | 8.46% | 100.00% | 2222 | 1.92079 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3053 | 0.87% | 5.55% | 57.10% | 963 | 1.37399 |
| anti | 2294 | 0.65% | 4.17% | 42.90% | 1290 | 1.0324 |
| both | 5347 | 1.52% | 9.72% | 100.00% | 2222 | 2.40639 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
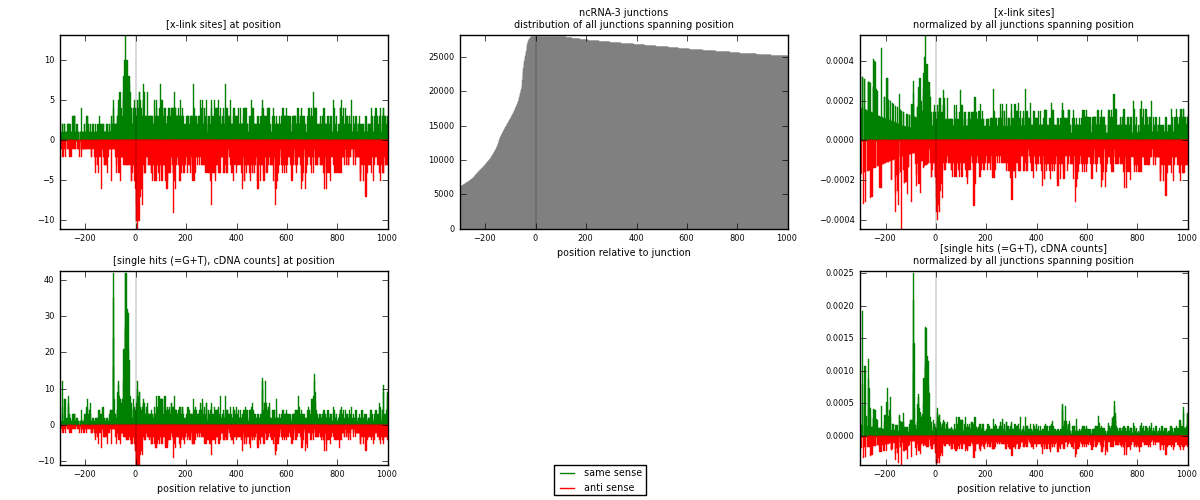 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2284 | 0.69% | 4.53% | 49.57% | 1040 | 0.965751 |
| anti | 2324 | 0.70% | 4.61% | 50.43% | 1349 | 0.982664 |
| both | 4608 | 1.39% | 9.14% | 100.00% | 2365 | 1.94841 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3229 | 0.92% | 5.87% | 56.70% | 1040 | 1.36533 |
| anti | 2466 | 0.70% | 4.48% | 43.30% | 1349 | 1.04271 |
| both | 5695 | 1.61% | 10.35% | 100.00% | 2365 | 2.40803 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
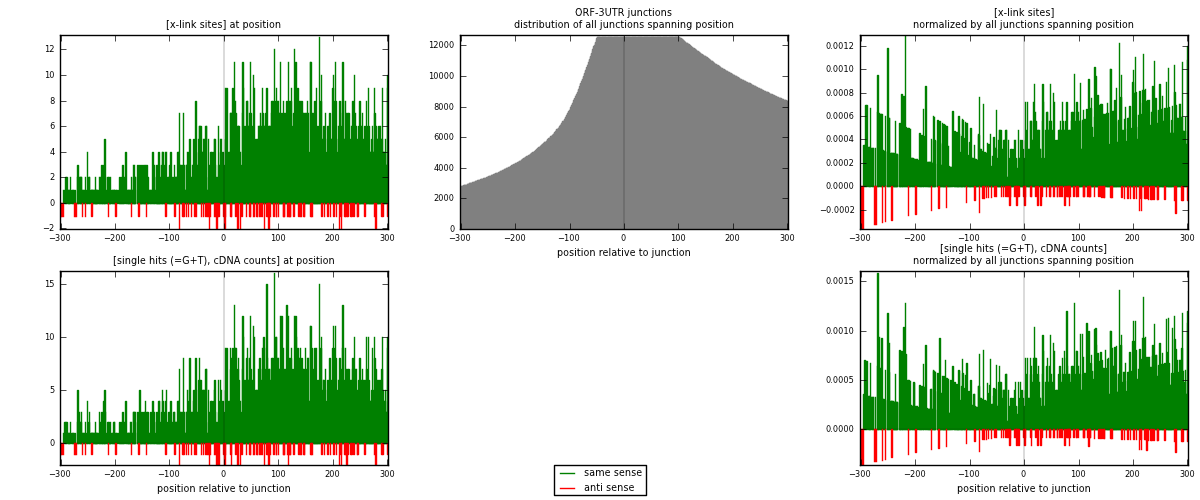 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2260 | 0.68% | 4.48% | 95.00% | 1264 | 1.66544 |
| anti | 119 | 0.04% | 0.24% | 5.00% | 103 | 0.0876934 |
| both | 2379 | 0.72% | 4.72% | 100.00% | 1357 | 1.75313 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2457 | 0.70% | 4.46% | 95.23% | 1264 | 1.81061 |
| anti | 123 | 0.03% | 0.22% | 4.77% | 103 | 0.0906411 |
| both | 2580 | 0.73% | 4.69% | 100.00% | 1357 | 1.90125 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
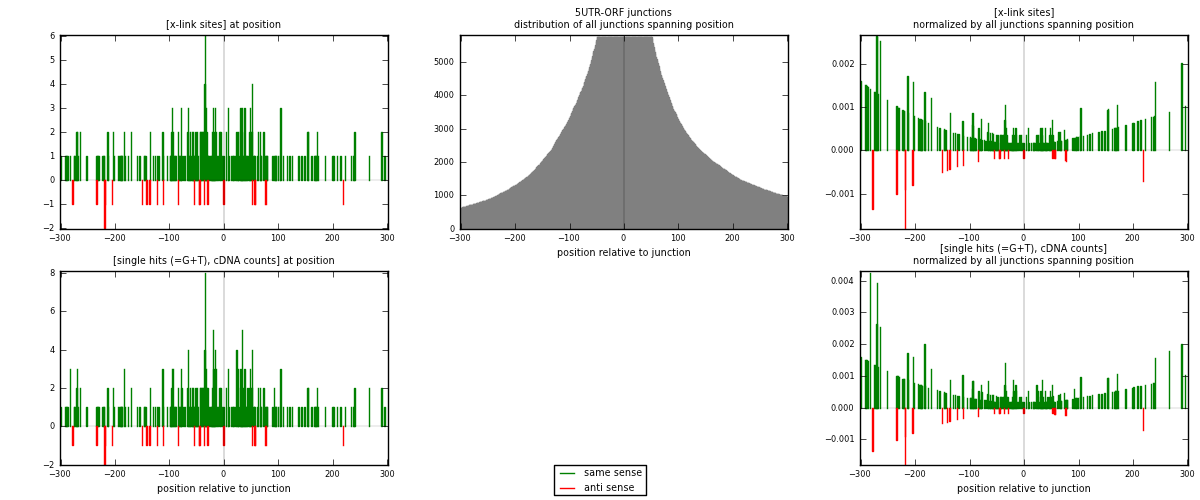 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 277 | 0.08% | 0.55% | 92.03% | 174 | 1.45026 |
| anti | 24 | 0.01% | 0.05% | 7.97% | 17 | 0.125654 |
| both | 301 | 0.09% | 0.60% | 100.00% | 191 | 1.57592 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 302 | 0.09% | 0.55% | 92.64% | 174 | 1.58115 |
| anti | 24 | 0.01% | 0.04% | 7.36% | 17 | 0.125654 |
| both | 326 | 0.09% | 0.59% | 100.00% | 191 | 1.70681 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
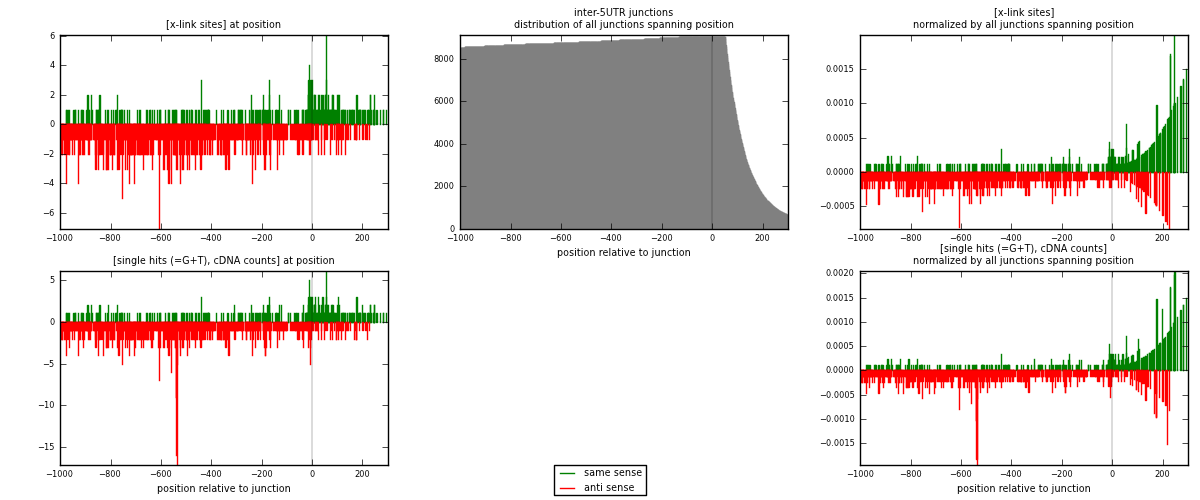 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 322 | 0.10% | 0.64% | 27.45% | 226 | 0.434548 |
| anti | 851 | 0.26% | 1.69% | 72.55% | 536 | 1.14845 |
| both | 1173 | 0.35% | 2.33% | 100.00% | 741 | 1.583 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 340 | 0.10% | 0.62% | 26.34% | 226 | 0.458839 |
| anti | 951 | 0.27% | 1.73% | 73.66% | 536 | 1.2834 |
| both | 1291 | 0.37% | 2.35% | 100.00% | 741 | 1.74224 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
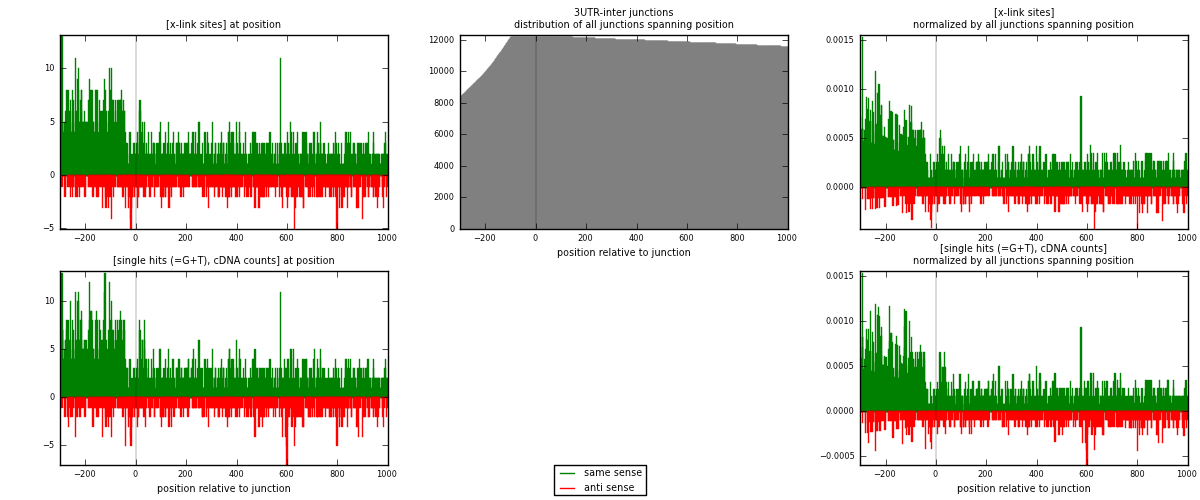 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2609 | 0.79% | 5.17% | 75.58% | 1583 | 1.28081 |
| anti | 843 | 0.25% | 1.67% | 24.42% | 515 | 0.413844 |
| both | 3452 | 1.04% | 6.84% | 100.00% | 2037 | 1.69465 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2788 | 0.79% | 5.07% | 75.60% | 1583 | 1.36868 |
| anti | 900 | 0.26% | 1.64% | 24.40% | 515 | 0.441826 |
| both | 3688 | 1.05% | 6.70% | 100.00% | 2037 | 1.81051 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
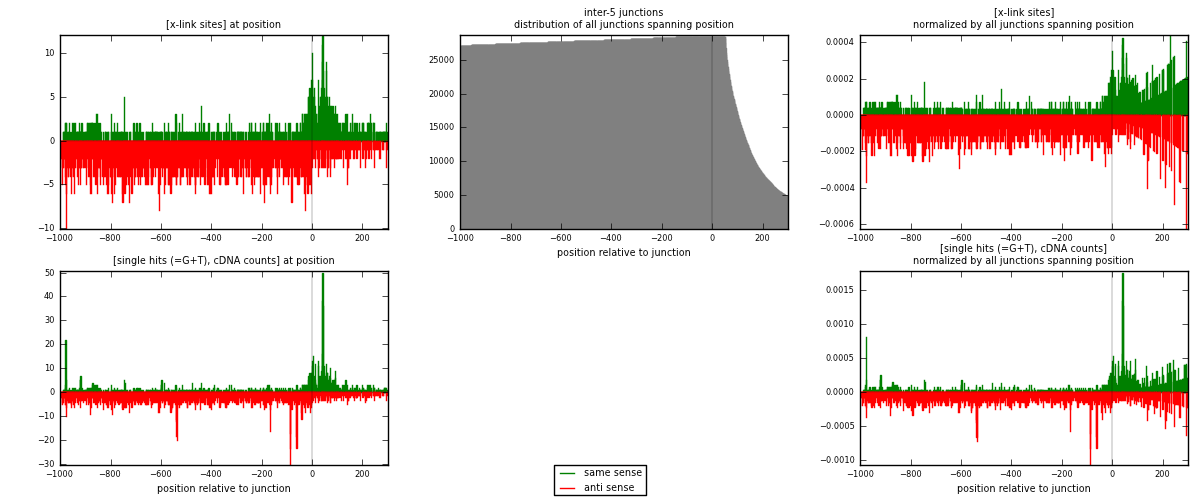
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1017 | 0.31% | 2.02% | 27.30% | 522 | 0.485442 |
| anti | 2708 | 0.82% | 5.37% | 72.70% | 1616 | 1.2926 |
| both | 3725 | 1.12% | 7.39% | 100.00% | 2095 | 1.77804 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1367 | 0.39% | 2.48% | 30.91% | 522 | 0.652506 |
| anti | 3056 | 0.87% | 5.55% | 69.09% | 1616 | 1.45871 |
| both | 4423 | 1.25% | 8.04% | 100.00% | 2095 | 2.11122 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
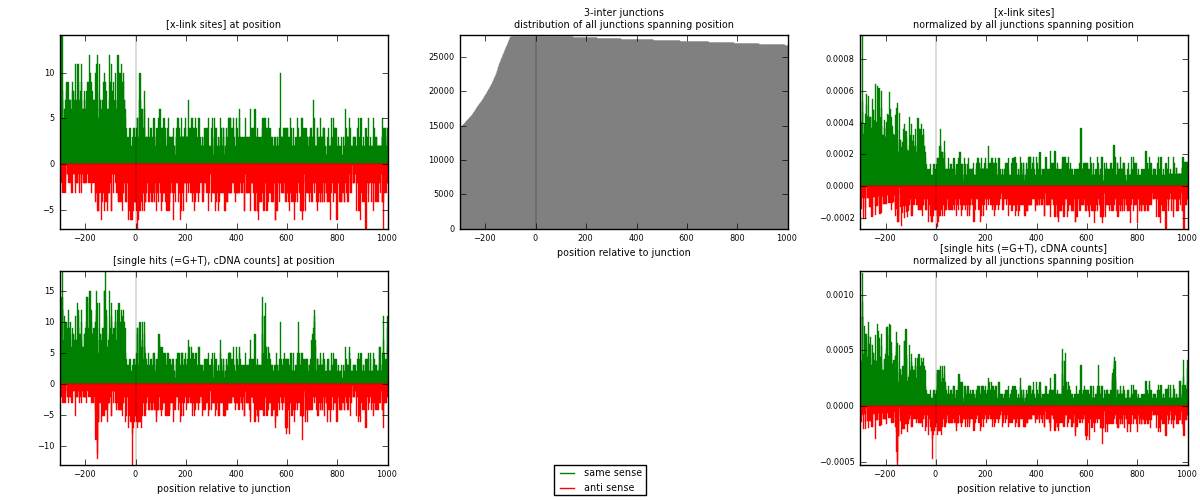
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3605 | 1.09% | 7.15% | 59.62% | 2006 | 1.09441 |
| anti | 2442 | 0.74% | 4.84% | 40.38% | 1370 | 0.741348 |
| both | 6047 | 1.82% | 11.99% | 100.00% | 3294 | 1.83576 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4247 | 1.20% | 7.72% | 61.44% | 2006 | 1.28931 |
| anti | 2665 | 0.76% | 4.84% | 38.56% | 1370 | 0.809047 |
| both | 6912 | 1.96% | 12.56% | 100.00% | 3294 | 2.09836 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
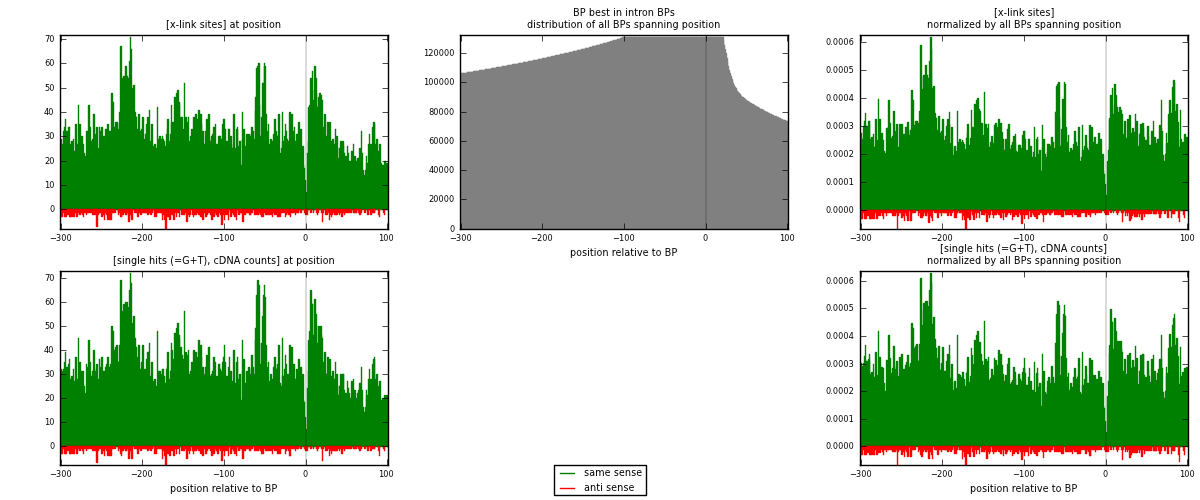
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 12568 | 3.79% | 24.92% | 95.80% | 9367 | 1.27854 |
| anti | 551 | 0.17% | 1.09% | 4.20% | 479 | 0.0560529 |
| both | 13119 | 3.95% | 26.01% | 100.00% | 9830 | 1.33459 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 13207 | 3.74% | 24.00% | 95.93% | 9367 | 1.34354 |
| anti | 560 | 0.16% | 1.02% | 4.07% | 479 | 0.0569685 |
| both | 13767 | 3.90% | 25.01% | 100.00% | 9830 | 1.40051 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.