RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
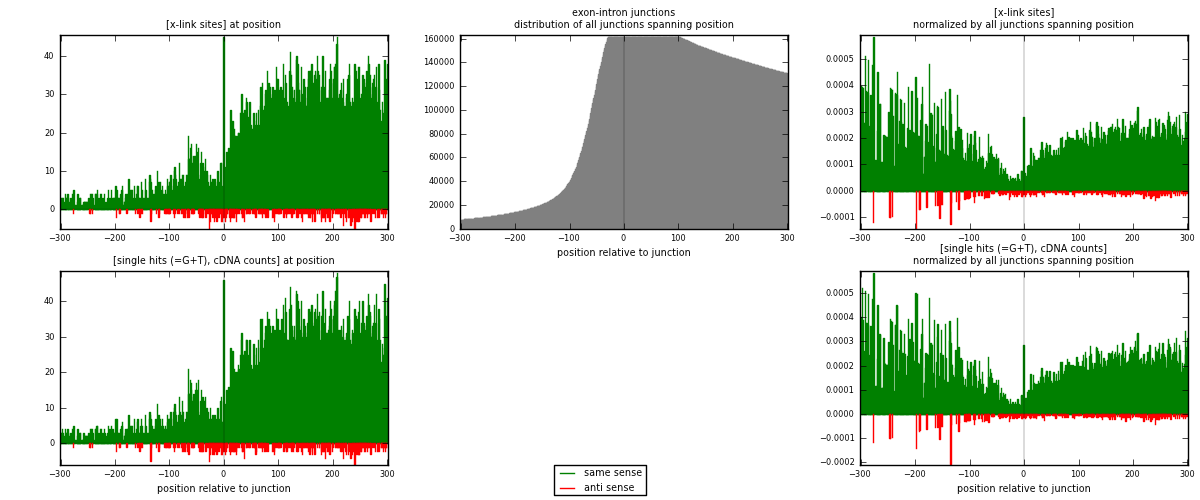 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9662 | 4.00% | 26.39% | 96.07% | 7556 | 1.22428 |
| anti | 395 | 0.16% | 1.08% | 3.93% | 356 | 0.0500507 |
| both | 10057 | 4.16% | 27.47% | 100.00% | 7892 | 1.27433 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 10150 | 3.95% | 25.32% | 96.07% | 7556 | 1.28611 |
| anti | 415 | 0.16% | 1.04% | 3.93% | 356 | 0.0525849 |
| both | 10565 | 4.11% | 26.35% | 100.00% | 7892 | 1.3387 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
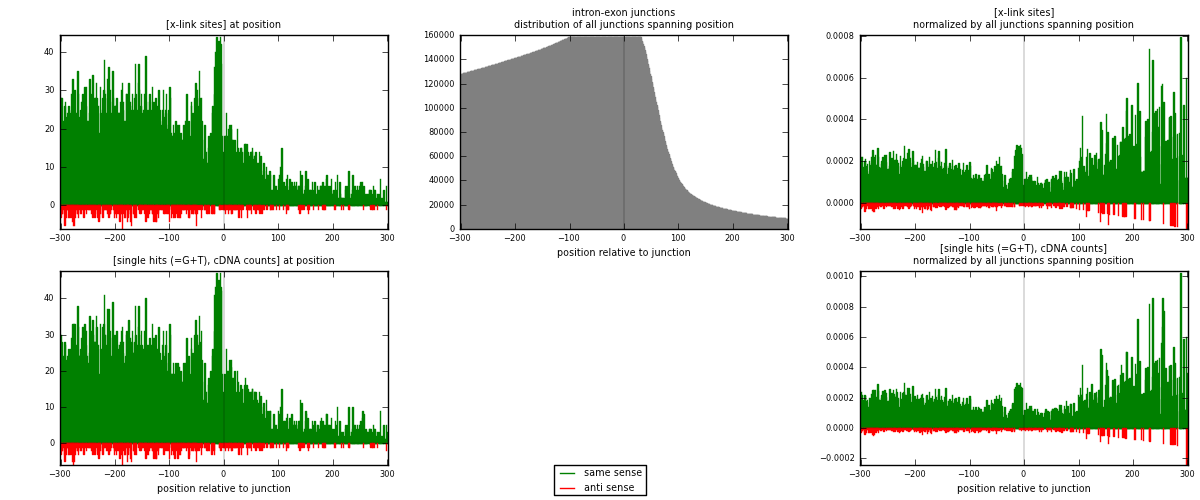 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8882 | 3.68% | 24.26% | 94.57% | 7002 | 1.19462 |
| anti | 510 | 0.21% | 1.39% | 5.43% | 446 | 0.0685945 |
| both | 9392 | 3.89% | 25.66% | 100.00% | 7435 | 1.26321 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 9330 | 3.63% | 23.27% | 94.63% | 7002 | 1.25488 |
| anti | 529 | 0.21% | 1.32% | 5.37% | 446 | 0.07115 |
| both | 9859 | 3.84% | 24.59% | 100.00% | 7435 | 1.32603 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
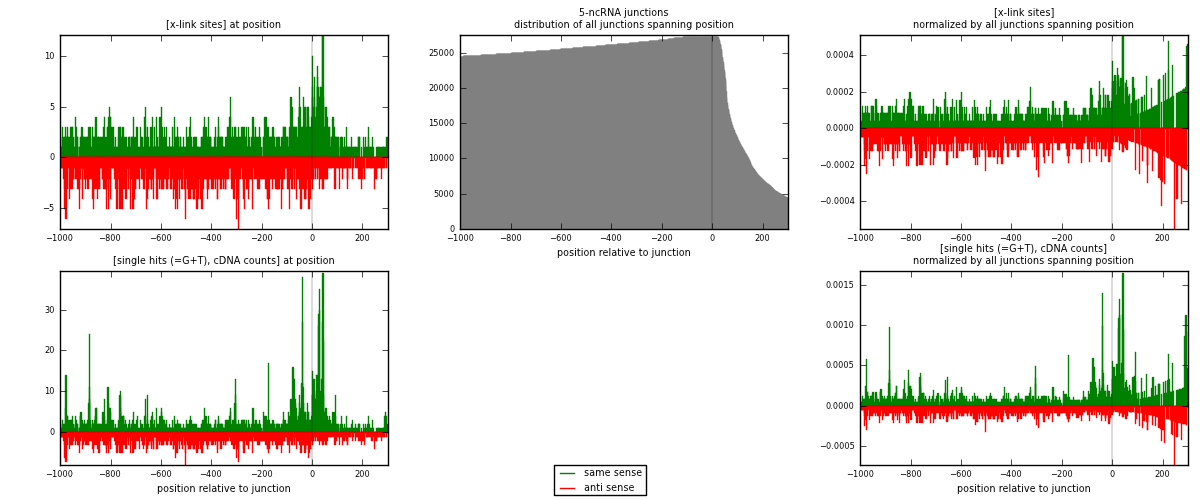 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1644 | 0.68% | 4.49% | 49.07% | 822 | 0.875866 |
| anti | 1706 | 0.71% | 4.66% | 50.93% | 1082 | 0.908897 |
| both | 3350 | 1.39% | 9.15% | 100.00% | 1877 | 1.78476 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2420 | 0.94% | 6.04% | 57.55% | 822 | 1.28929 |
| anti | 1785 | 0.69% | 4.45% | 42.45% | 1082 | 0.950986 |
| both | 4205 | 1.64% | 10.49% | 100.00% | 1877 | 2.24028 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
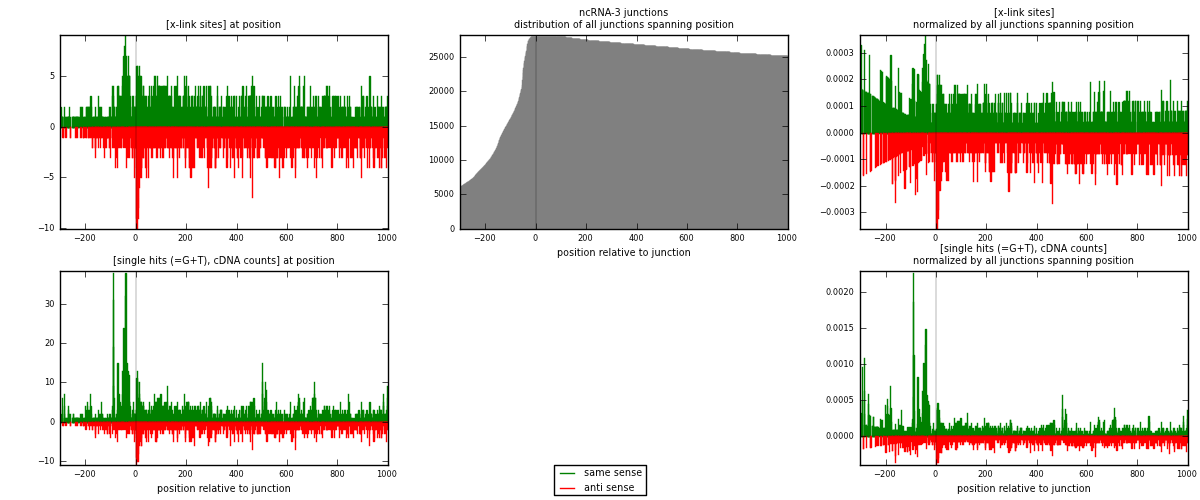 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1874 | 0.78% | 5.12% | 53.03% | 887 | 0.974012 |
| anti | 1660 | 0.69% | 4.53% | 46.97% | 1052 | 0.862786 |
| both | 3534 | 1.46% | 9.65% | 100.00% | 1924 | 1.8368 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 2599 | 1.01% | 6.48% | 59.76% | 887 | 1.35083 |
| anti | 1750 | 0.68% | 4.36% | 40.24% | 1052 | 0.909563 |
| both | 4349 | 1.69% | 10.85% | 100.00% | 1924 | 2.2604 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
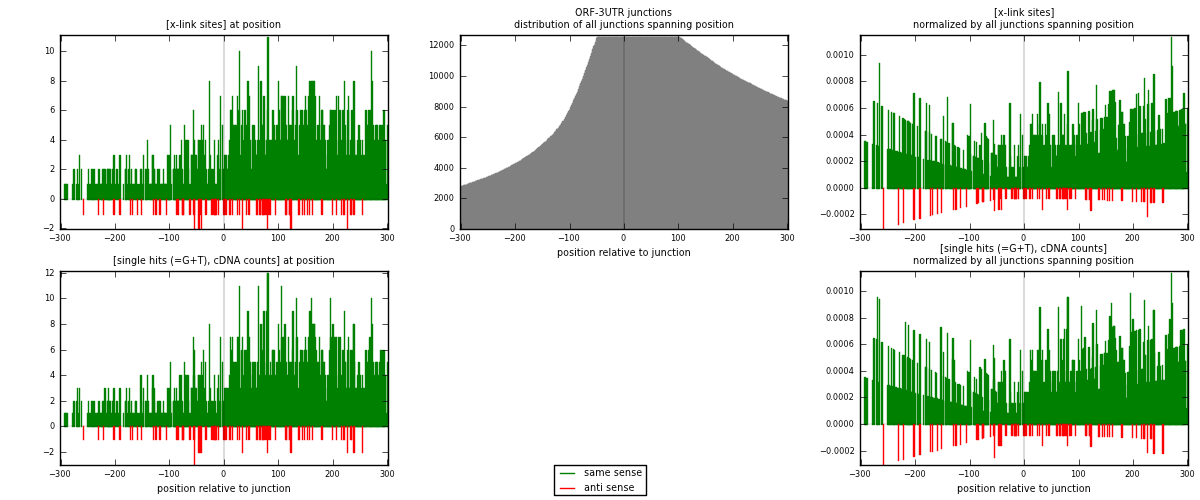
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1576 | 0.65% | 4.31% | 94.83% | 970 | 1.52418 |
| anti | 86 | 0.04% | 0.23% | 5.17% | 72 | 0.0831721 |
| both | 1662 | 0.69% | 4.54% | 100.00% | 1034 | 1.60735 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1695 | 0.66% | 4.23% | 94.96% | 970 | 1.63926 |
| anti | 90 | 0.04% | 0.22% | 5.04% | 72 | 0.0870406 |
| both | 1785 | 0.69% | 4.45% | 100.00% | 1034 | 1.72631 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
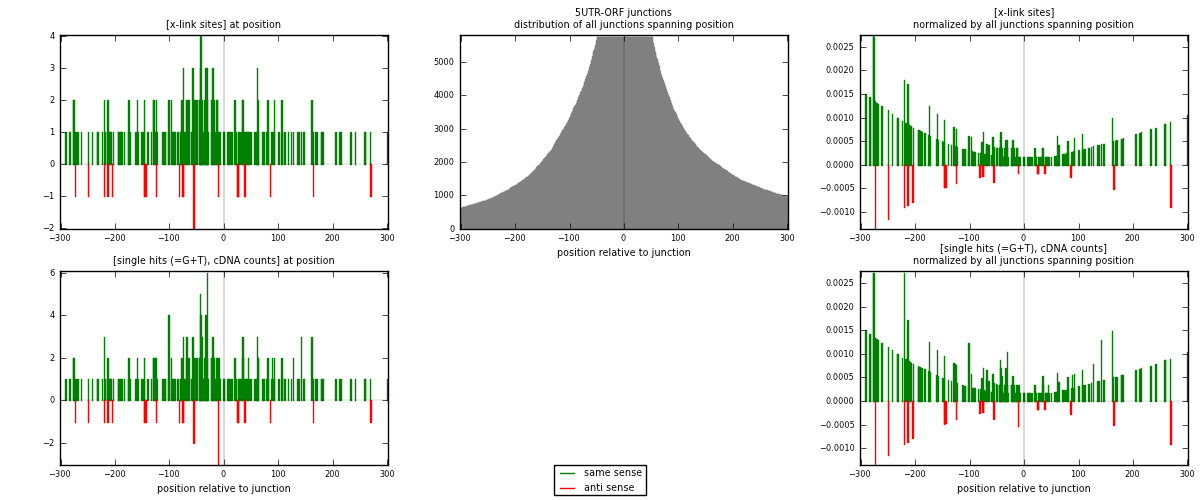
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 231 | 0.10% | 0.63% | 90.94% | 160 | 1.3125 |
| anti | 23 | 0.01% | 0.06% | 9.06% | 16 | 0.130682 |
| both | 254 | 0.11% | 0.69% | 100.00% | 176 | 1.44318 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 255 | 0.10% | 0.64% | 91.07% | 160 | 1.44886 |
| anti | 25 | 0.01% | 0.06% | 8.93% | 16 | 0.142045 |
| both | 280 | 0.11% | 0.70% | 100.00% | 176 | 1.59091 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
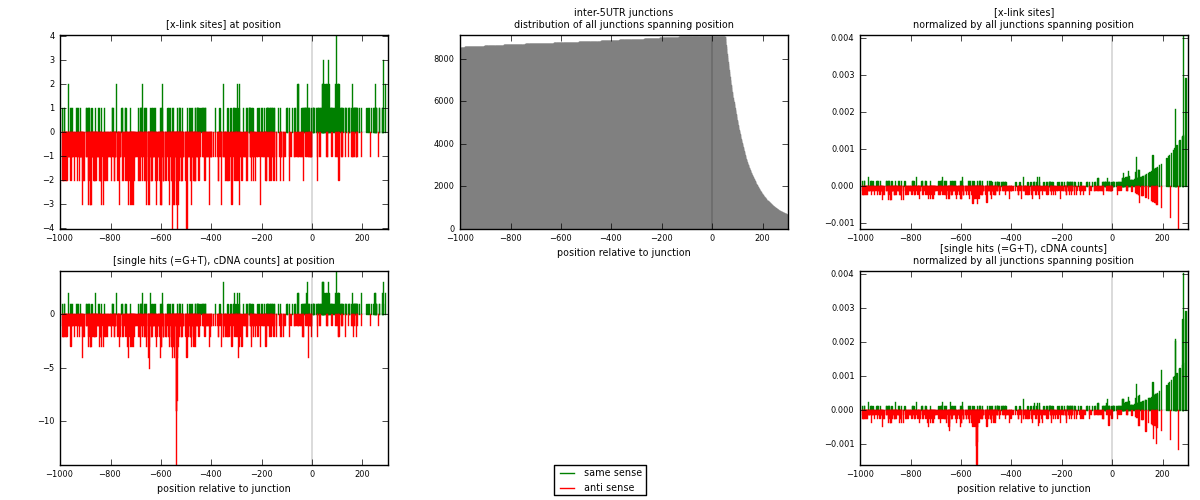
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 263 | 0.11% | 0.72% | 28.34% | 178 | 0.445008 |
| anti | 665 | 0.28% | 1.82% | 71.66% | 426 | 1.12521 |
| both | 928 | 0.38% | 2.54% | 100.00% | 591 | 1.57022 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 274 | 0.11% | 0.68% | 26.73% | 178 | 0.463621 |
| anti | 751 | 0.29% | 1.87% | 73.27% | 426 | 1.27073 |
| both | 1025 | 0.40% | 2.56% | 100.00% | 591 | 1.73435 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
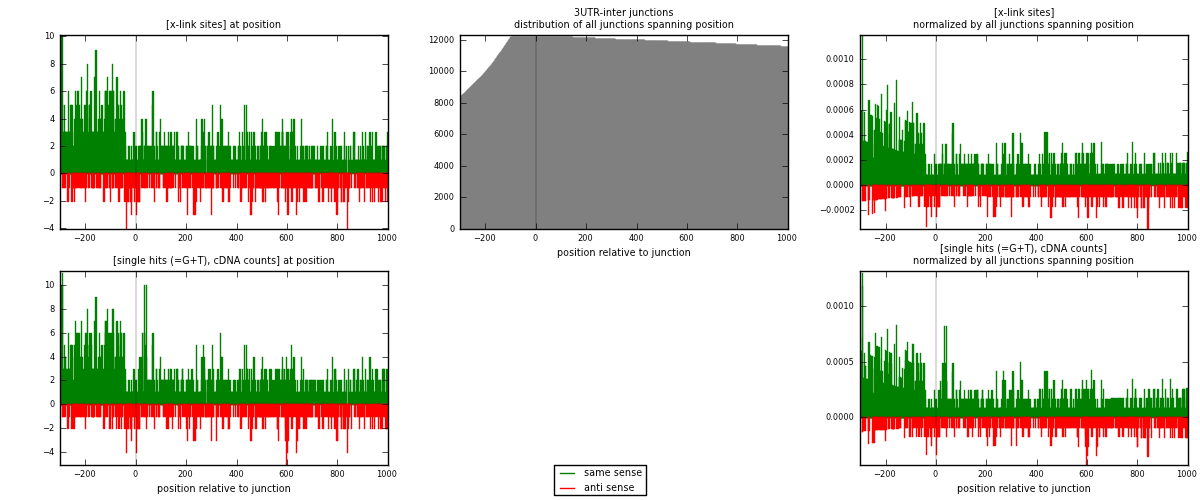
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1829 | 0.76% | 5.00% | 76.46% | 1175 | 1.19621 |
| anti | 563 | 0.23% | 1.54% | 23.54% | 383 | 0.368215 |
| both | 2392 | 0.99% | 6.53% | 100.00% | 1529 | 1.56442 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1975 | 0.77% | 4.93% | 76.61% | 1175 | 1.29169 |
| anti | 603 | 0.23% | 1.50% | 23.39% | 383 | 0.394375 |
| both | 2578 | 1.00% | 6.43% | 100.00% | 1529 | 1.68607 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
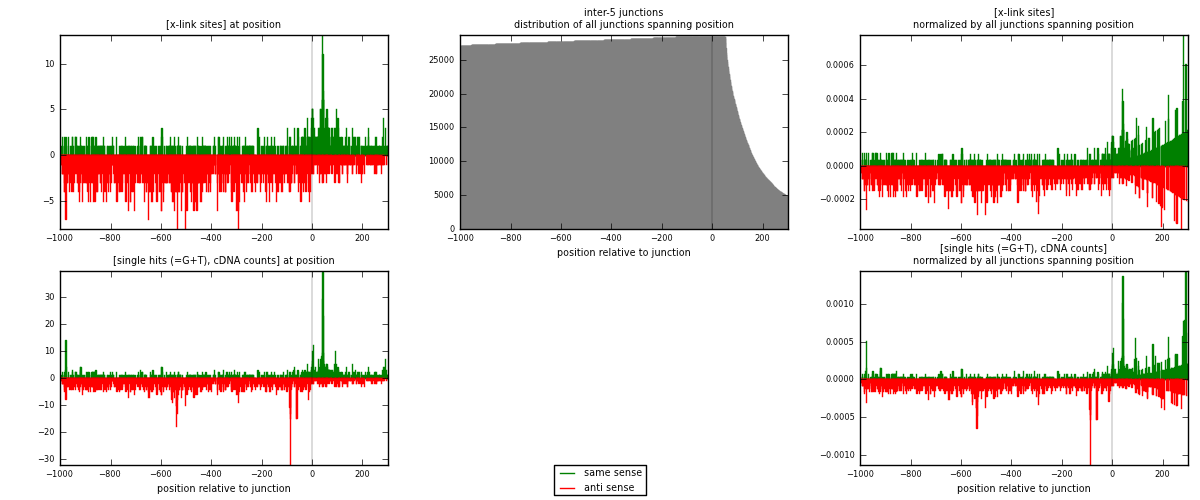 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 800 | 0.33% | 2.19% | 27.23% | 448 | 0.460035 |
| anti | 2138 | 0.89% | 5.84% | 72.77% | 1327 | 1.22944 |
| both | 2938 | 1.22% | 8.03% | 100.00% | 1739 | 1.68948 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1005 | 0.39% | 2.51% | 29.45% | 448 | 0.577918 |
| anti | 2408 | 0.94% | 6.01% | 70.55% | 1327 | 1.3847 |
| both | 3413 | 1.33% | 8.51% | 100.00% | 1739 | 1.96262 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
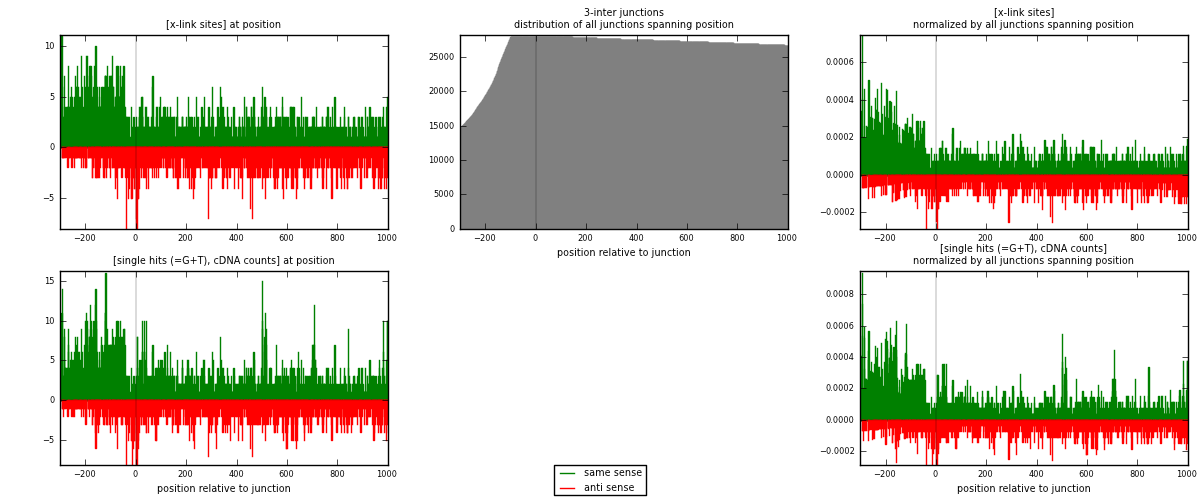 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2631 | 1.09% | 7.19% | 60.40% | 1525 | 1.03176 |
| anti | 1725 | 0.71% | 4.71% | 39.60% | 1066 | 0.676471 |
| both | 4356 | 1.80% | 11.90% | 100.00% | 2550 | 1.70824 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 3185 | 1.24% | 7.94% | 63.14% | 1525 | 1.24902 |
| anti | 1859 | 0.72% | 4.64% | 36.86% | 1066 | 0.72902 |
| both | 5044 | 1.96% | 12.58% | 100.00% | 2550 | 1.97804 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
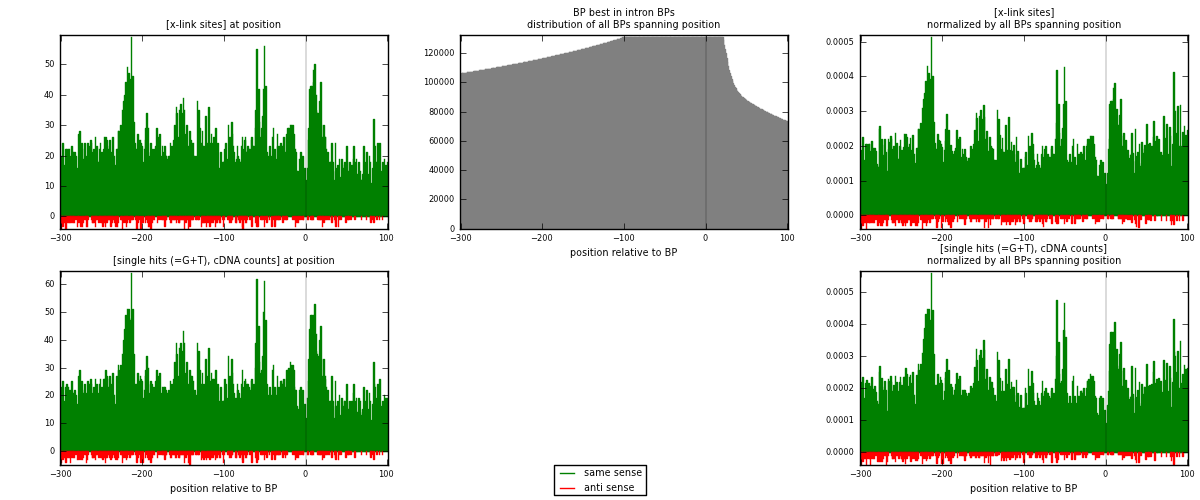 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 9175 | 3.80% | 25.06% | 95.62% | 7168 | 1.21797 |
| anti | 420 | 0.17% | 1.15% | 4.38% | 381 | 0.0557547 |
| both | 9595 | 3.97% | 26.21% | 100.00% | 7533 | 1.27373 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 9640 | 3.75% | 24.04% | 95.65% | 7168 | 1.2797 |
| anti | 438 | 0.17% | 1.09% | 4.35% | 381 | 0.0581442 |
| both | 10078 | 3.92% | 25.14% | 100.00% | 7533 | 1.33785 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.