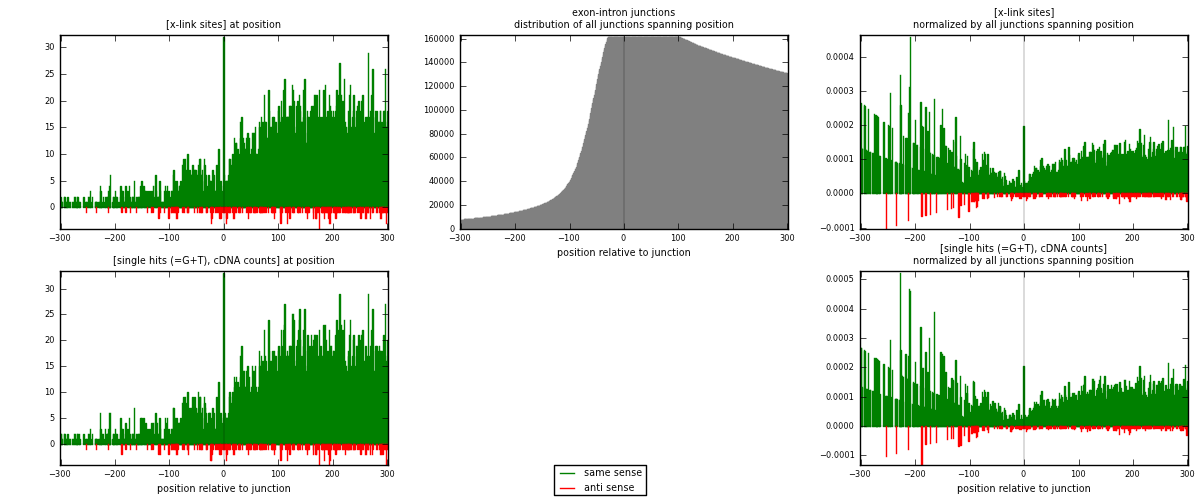
RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5151 | 3.97% | 25.97% | 95.73% | 4317 | 1.14238 |
| anti | 230 | 0.18% | 1.16% | 4.27% | 195 | 0.0510091 |
| both | 5381 | 4.14% | 27.13% | 100.00% | 4509 | 1.19339 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 5401 | 3.94% | 25.11% | 95.73% | 4317 | 1.19783 |
| anti | 241 | 0.18% | 1.12% | 4.27% | 195 | 0.0534487 |
| both | 5642 | 4.11% | 26.23% | 100.00% | 4509 | 1.25128 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
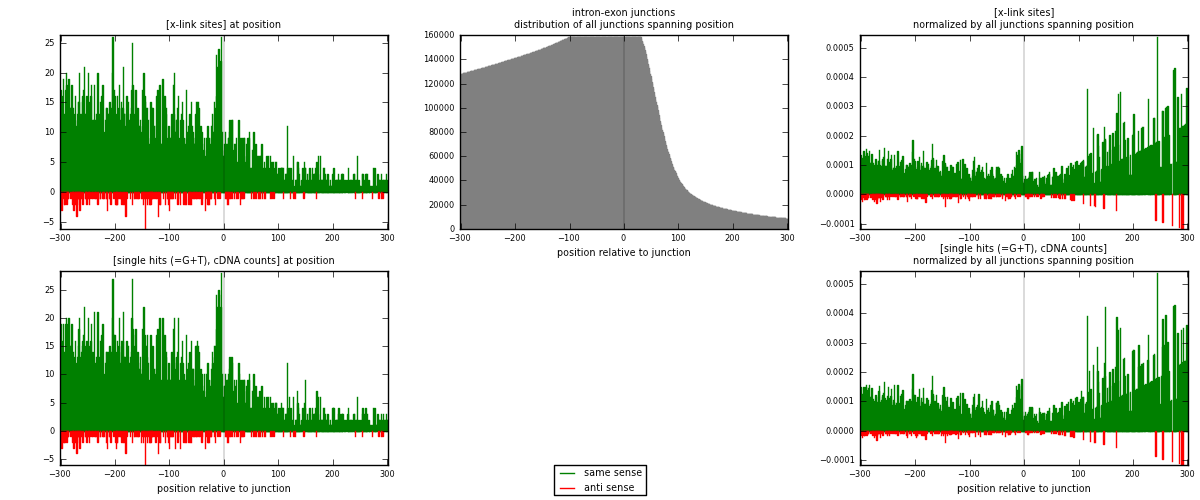
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4574 | 3.52% | 23.06% | 94.52% | 3900 | 1.10831 |
| anti | 265 | 0.20% | 1.34% | 5.48% | 235 | 0.0642113 |
| both | 4839 | 3.73% | 24.40% | 100.00% | 4127 | 1.17252 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 4773 | 3.48% | 22.19% | 94.59% | 3900 | 1.15653 |
| anti | 273 | 0.20% | 1.27% | 5.41% | 235 | 0.0661497 |
| both | 5046 | 3.68% | 23.46% | 100.00% | 4127 | 1.22268 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
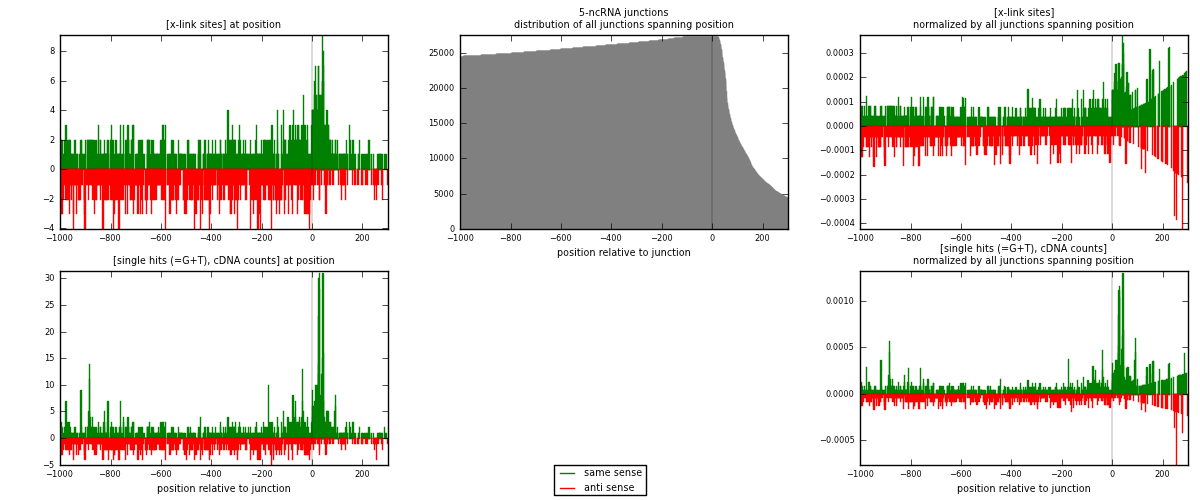
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1032 | 0.79% | 5.20% | 52.90% | 538 | 0.848684 |
| anti | 919 | 0.71% | 4.63% | 47.10% | 687 | 0.755757 |
| both | 1951 | 1.50% | 9.84% | 100.00% | 1216 | 1.60444 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1432 | 1.04% | 6.66% | 59.64% | 538 | 1.17763 |
| anti | 969 | 0.71% | 4.50% | 40.36% | 687 | 0.796875 |
| both | 2401 | 1.75% | 11.16% | 100.00% | 1216 | 1.97451 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
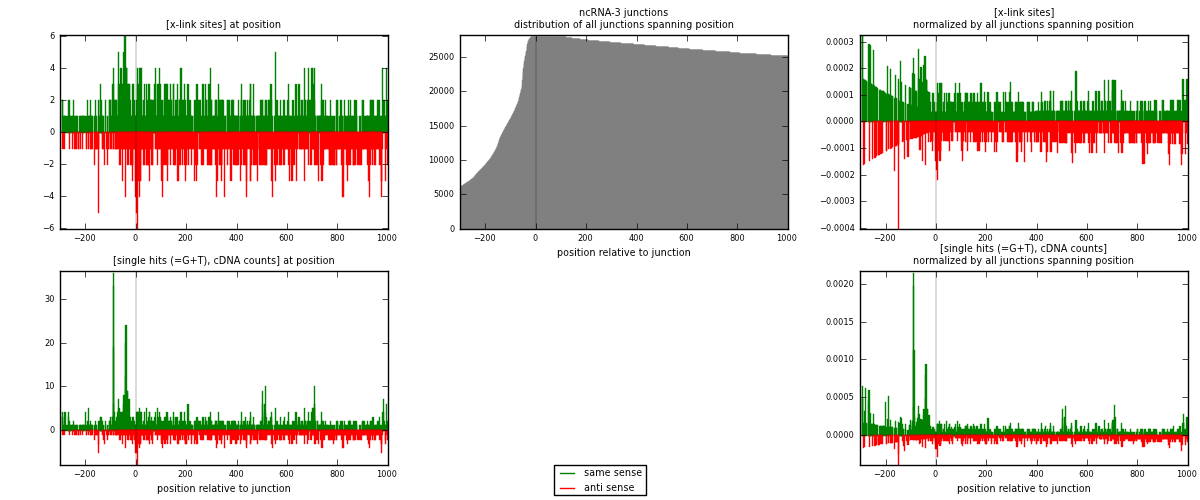
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1066 | 0.82% | 5.38% | 53.70% | 531 | 0.883914 |
| anti | 919 | 0.71% | 4.63% | 46.30% | 681 | 0.762023 |
| both | 1985 | 1.53% | 10.01% | 100.00% | 1206 | 1.64594 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1422 | 1.04% | 6.61% | 59.82% | 531 | 1.1791 |
| anti | 955 | 0.70% | 4.44% | 40.18% | 681 | 0.791874 |
| both | 2377 | 1.73% | 11.05% | 100.00% | 1206 | 1.97098 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
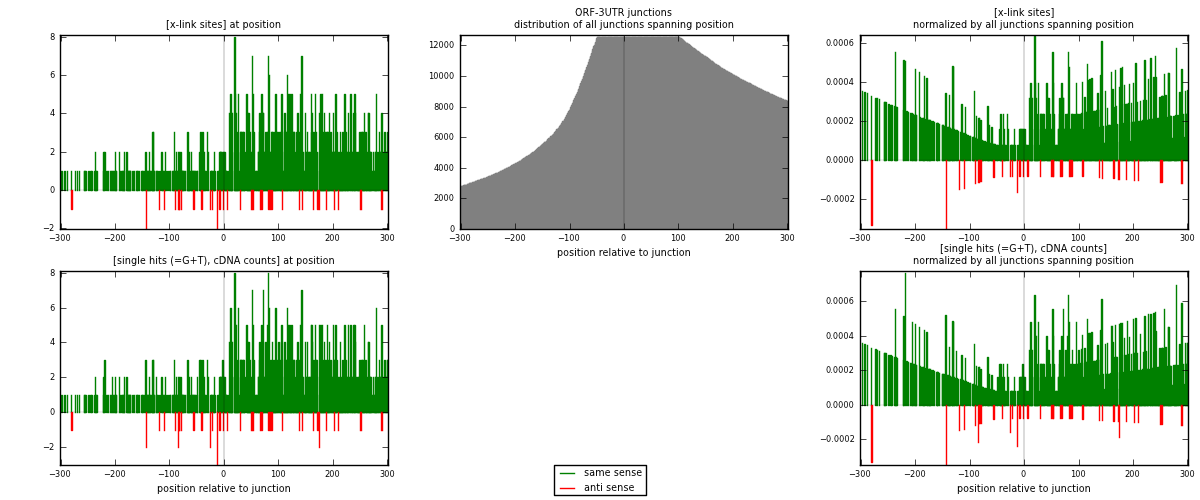
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 835 | 0.64% | 4.21% | 95.32% | 598 | 1.31496 |
| anti | 41 | 0.03% | 0.21% | 4.68% | 37 | 0.0645669 |
| both | 876 | 0.67% | 4.42% | 100.00% | 635 | 1.37953 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 882 | 0.64% | 4.10% | 95.15% | 598 | 1.38898 |
| anti | 45 | 0.03% | 0.21% | 4.85% | 37 | 0.0708661 |
| both | 927 | 0.68% | 4.31% | 100.00% | 635 | 1.45984 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
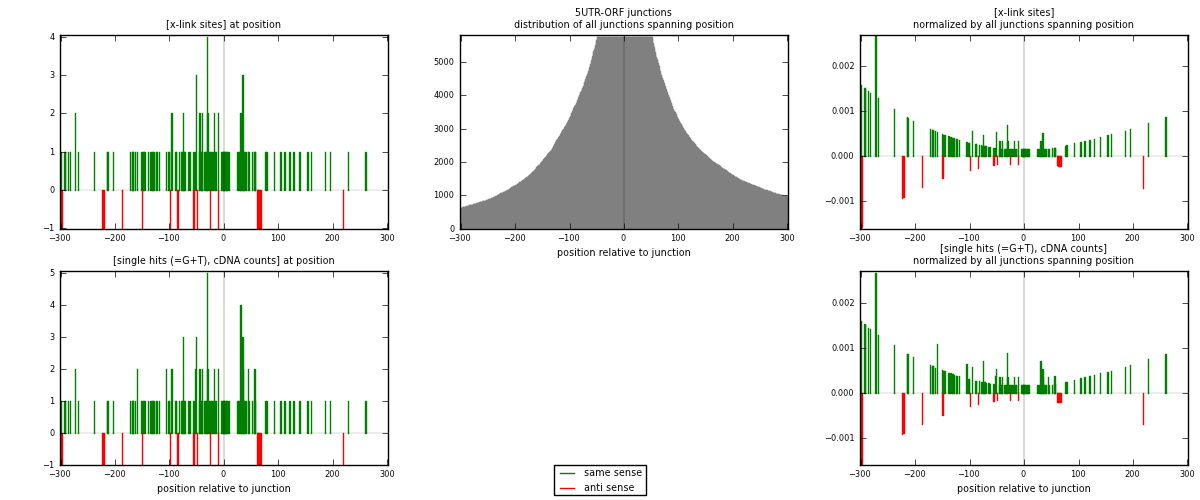
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 121 | 0.09% | 0.61% | 87.05% | 93 | 1.13084 |
| anti | 18 | 0.01% | 0.09% | 12.95% | 14 | 0.168224 |
| both | 139 | 0.11% | 0.70% | 100.00% | 107 | 1.29907 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 131 | 0.10% | 0.61% | 87.92% | 93 | 1.2243 |
| anti | 18 | 0.01% | 0.08% | 12.08% | 14 | 0.168224 |
| both | 149 | 0.11% | 0.69% | 100.00% | 107 | 1.39252 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
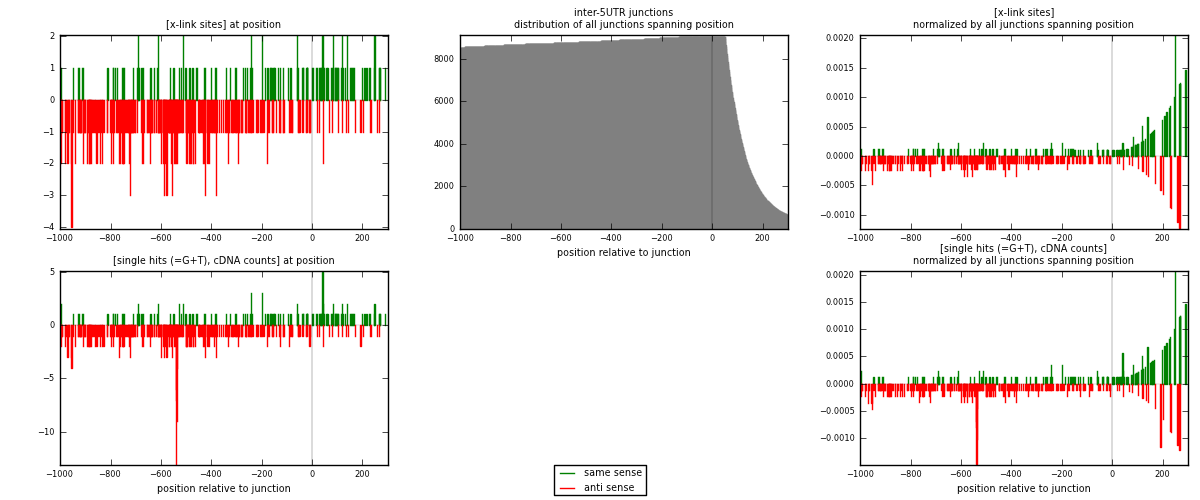
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 131 | 0.10% | 0.66% | 26.41% | 109 | 0.3531 |
| anti | 365 | 0.28% | 1.84% | 73.59% | 264 | 0.983827 |
| both | 496 | 0.38% | 2.50% | 100.00% | 371 | 1.33693 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 139 | 0.10% | 0.65% | 25.09% | 109 | 0.374663 |
| anti | 415 | 0.30% | 1.93% | 74.91% | 264 | 1.1186 |
| both | 554 | 0.40% | 2.58% | 100.00% | 371 | 1.49326 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
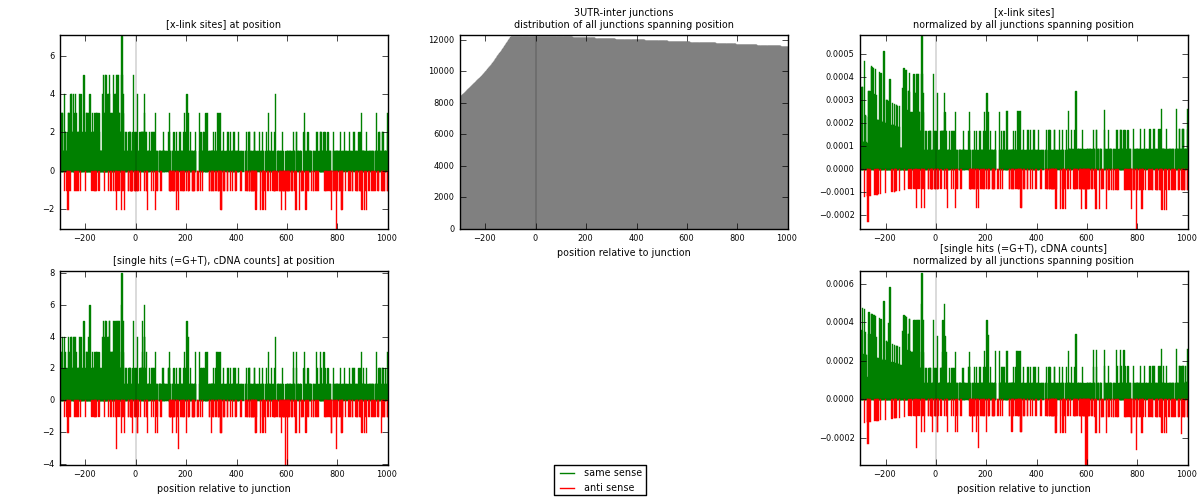
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1037 | 0.80% | 5.23% | 77.56% | 764 | 1.05708 |
| anti | 300 | 0.23% | 1.51% | 22.44% | 229 | 0.30581 |
| both | 1337 | 1.03% | 6.74% | 100.00% | 981 | 1.3629 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1099 | 0.80% | 5.11% | 77.56% | 764 | 1.12029 |
| anti | 318 | 0.23% | 1.48% | 22.44% | 229 | 0.324159 |
| both | 1417 | 1.03% | 6.59% | 100.00% | 981 | 1.44444 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
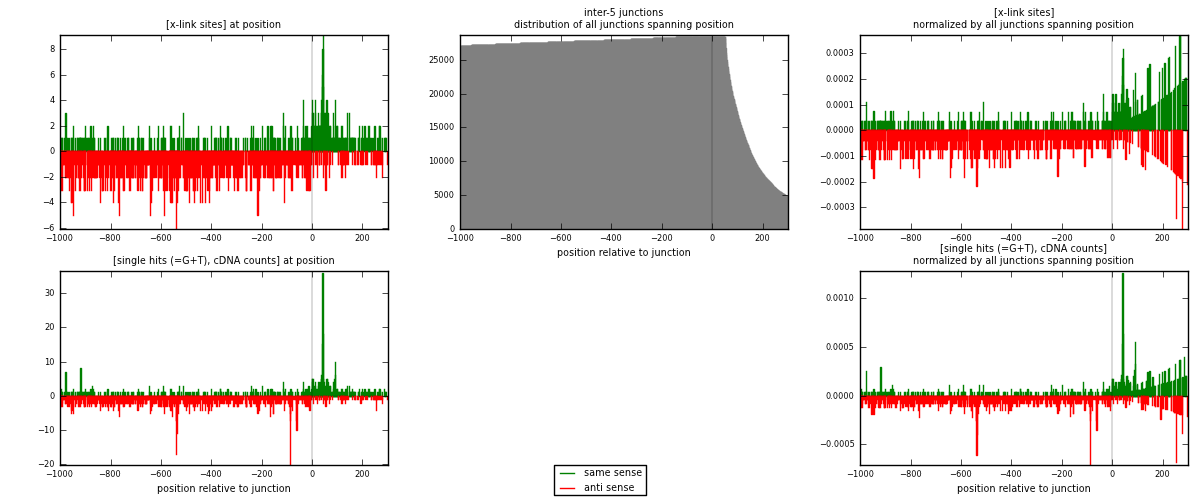
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 498 | 0.38% | 2.51% | 29.91% | 279 | 0.450271 |
| anti | 1167 | 0.90% | 5.88% | 70.09% | 835 | 1.05515 |
| both | 1665 | 1.28% | 8.40% | 100.00% | 1106 | 1.50542 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 627 | 0.46% | 2.91% | 32.27% | 279 | 0.566908 |
| anti | 1316 | 0.96% | 6.12% | 67.73% | 835 | 1.18987 |
| both | 1943 | 1.42% | 9.03% | 100.00% | 1106 | 1.75678 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
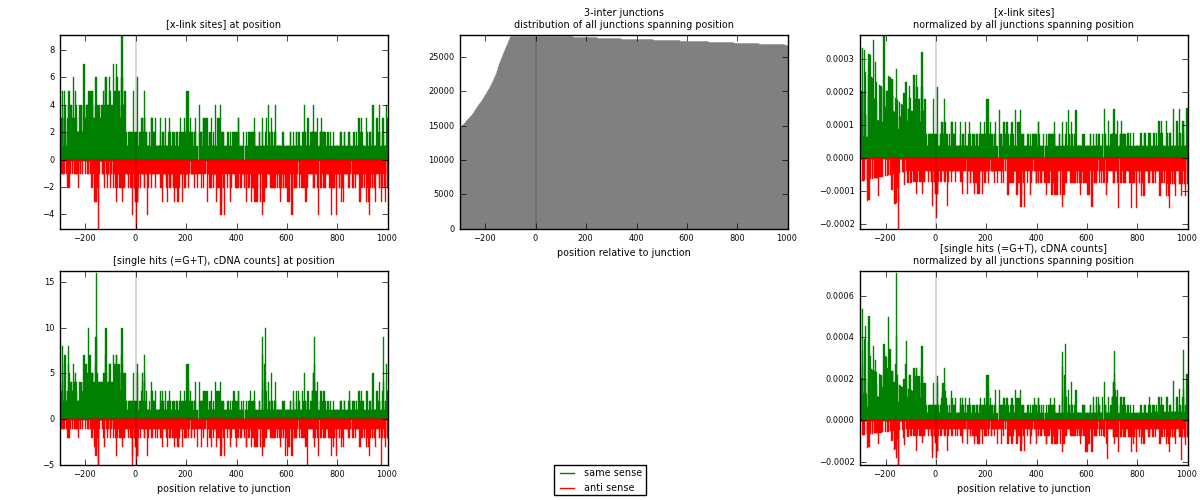
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1526 | 1.17% | 7.69% | 61.86% | 974 | 0.943723 |
| anti | 941 | 0.72% | 4.74% | 38.14% | 662 | 0.581942 |
| both | 2467 | 1.90% | 12.44% | 100.00% | 1617 | 1.52566 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [junctions] in map | [single hits (=G+T), cDNA counts] per unique [junction] |
| same | 1757 | 1.28% | 8.17% | 64.01% | 974 | 1.08658 |
| anti | 988 | 0.72% | 4.59% | 35.99% | 662 | 0.611008 |
| both | 2745 | 2.00% | 12.76% | 100.00% | 1617 | 1.69759 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
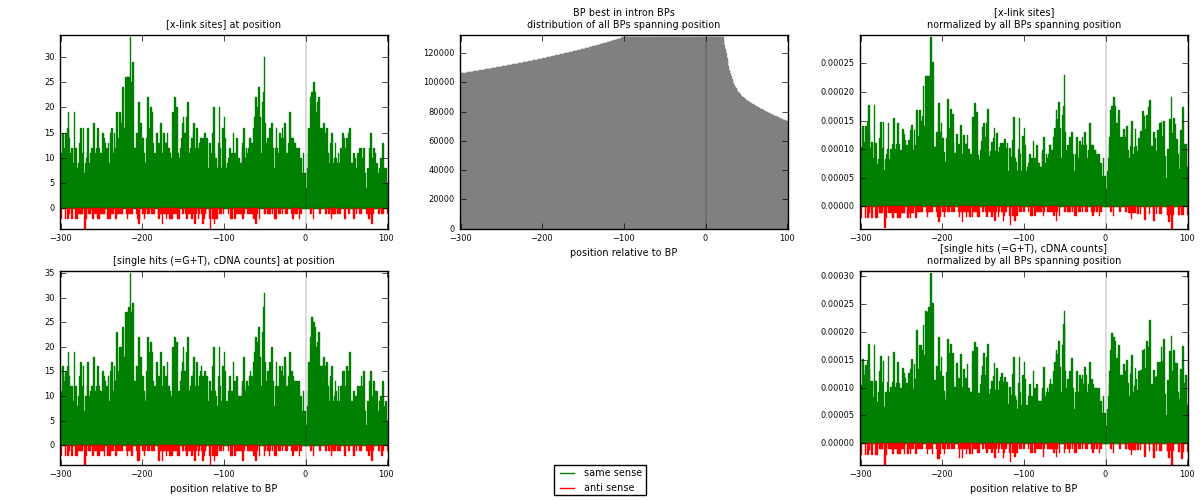
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 4931 | 3.80% | 24.86% | 95.05% | 4116 | 1.13435 |
| anti | 257 | 0.20% | 1.30% | 4.95% | 237 | 0.0591212 |
| both | 5188 | 3.99% | 26.16% | 100.00% | 4347 | 1.19347 |
| strand | [single hits (=G+T), cDNA counts] used in map | [single hits (=G+T), cDNA counts] % of total | [single hits (=G+T), cDNA counts] % of used in any map | [single hits (=G+T), cDNA counts] % of used in map | unique [BPs] in map | [single hits (=G+T), cDNA counts] per unique [BP] |
| same | 5148 | 3.75% | 23.93% | 95.10% | 4116 | 1.18427 |
| anti | 265 | 0.19% | 1.23% | 4.90% | 237 | 0.0609616 |
| both | 5413 | 3.95% | 25.16% | 100.00% | 4347 | 1.24523 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits (=G+T), cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.