RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
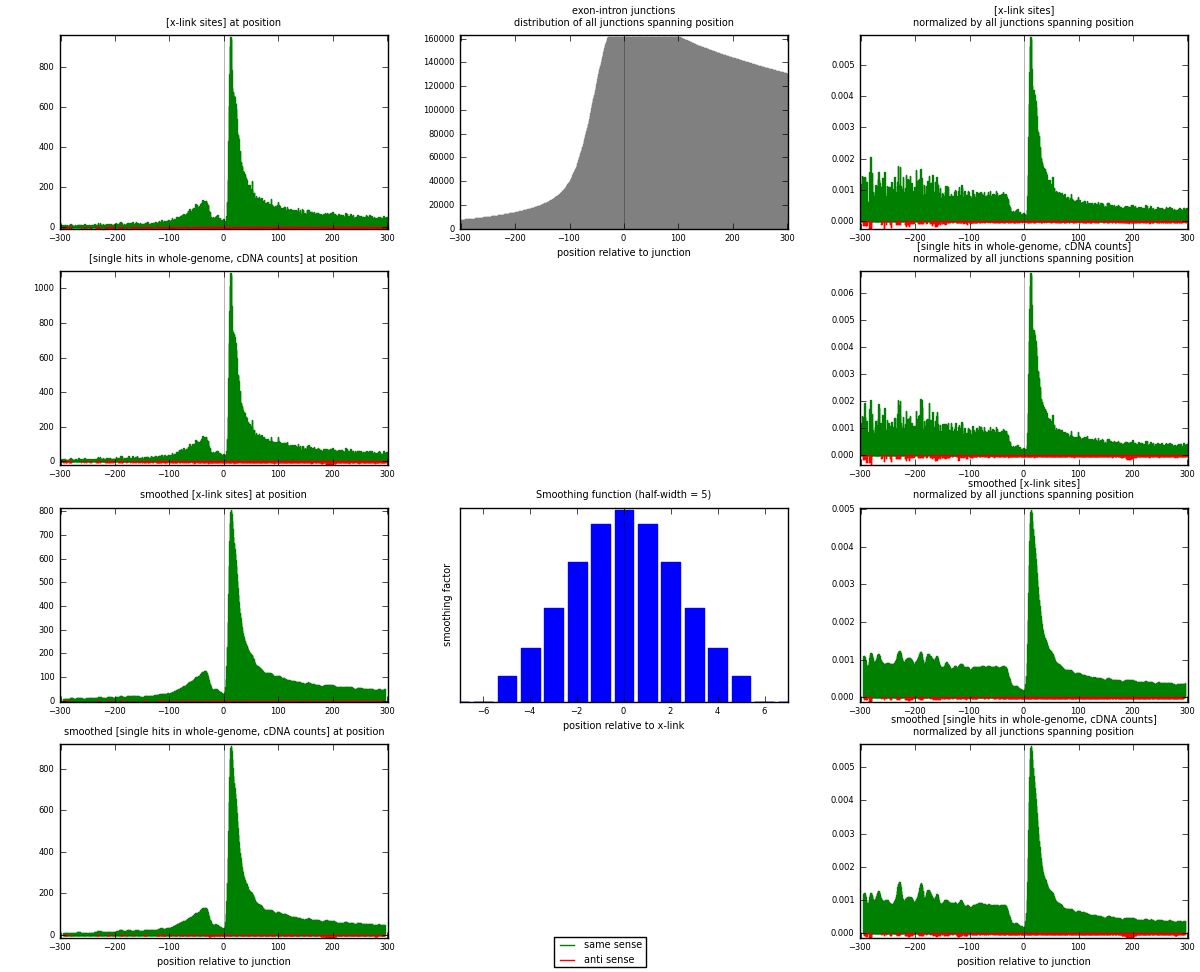 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 48203 | 11.15% | 35.31% | 98.06% | 28637 | 1.64971 |
| anti | 952 | 0.22% | 0.70% | 1.94% | 693 | 0.0325815 |
| both | 49155 | 11.37% | 36.01% | 100.00% | 29219 | 1.6823 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 51315 | 10.81% | 33.03% | 97.70% | 28637 | 1.75622 |
| anti | 1208 | 0.25% | 0.78% | 2.30% | 693 | 0.041343 |
| both | 52523 | 11.06% | 33.81% | 100.00% | 29219 | 1.79756 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
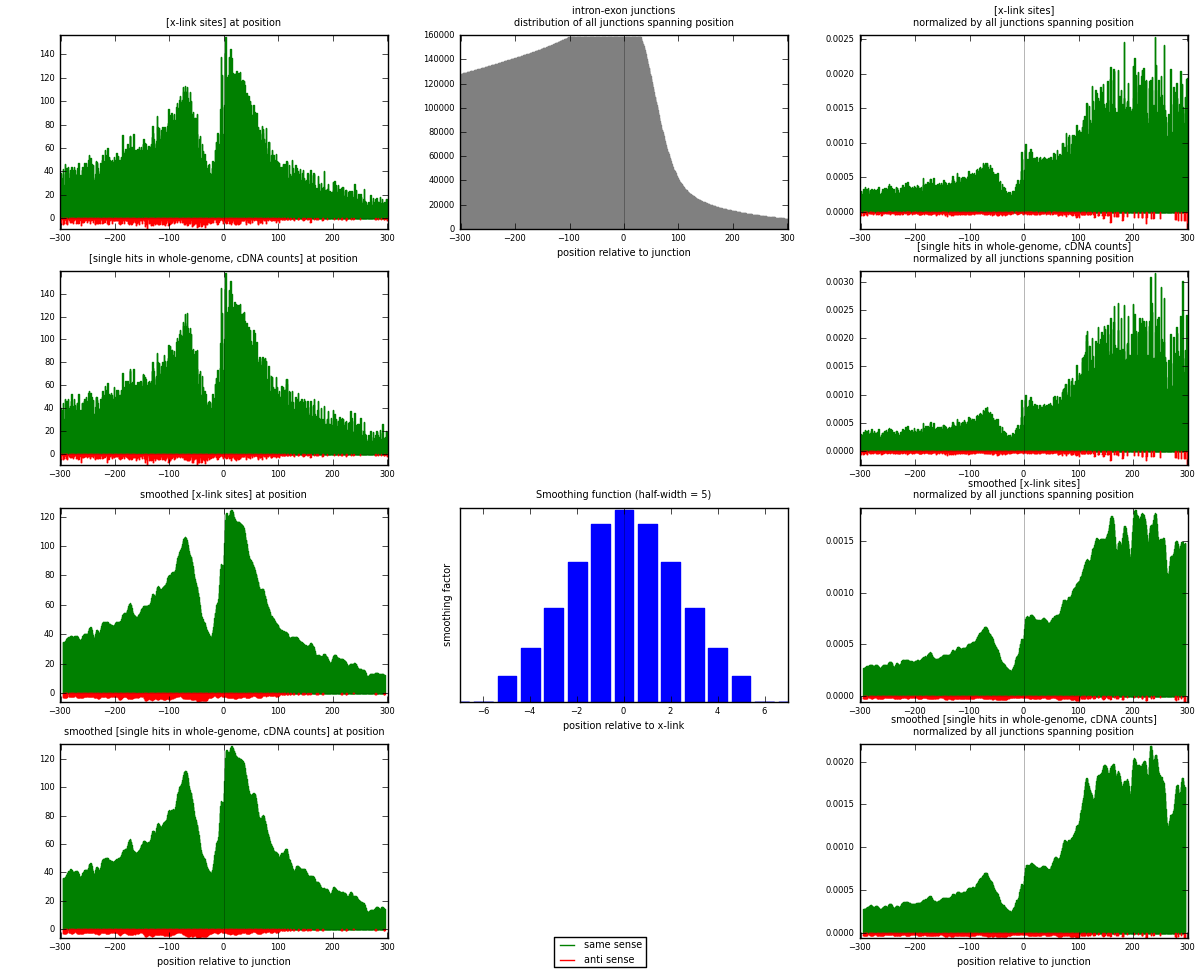 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 31396 | 7.26% | 23.00% | 96.87% | 20529 | 1.48094 |
| anti | 1016 | 0.23% | 0.74% | 3.13% | 762 | 0.0479245 |
| both | 32412 | 7.50% | 23.74% | 100.00% | 21200 | 1.52887 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 33579 | 7.07% | 21.62% | 96.91% | 20529 | 1.58392 |
| anti | 1069 | 0.23% | 0.69% | 3.09% | 762 | 0.0504245 |
| both | 34648 | 7.30% | 22.30% | 100.00% | 21200 | 1.63434 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
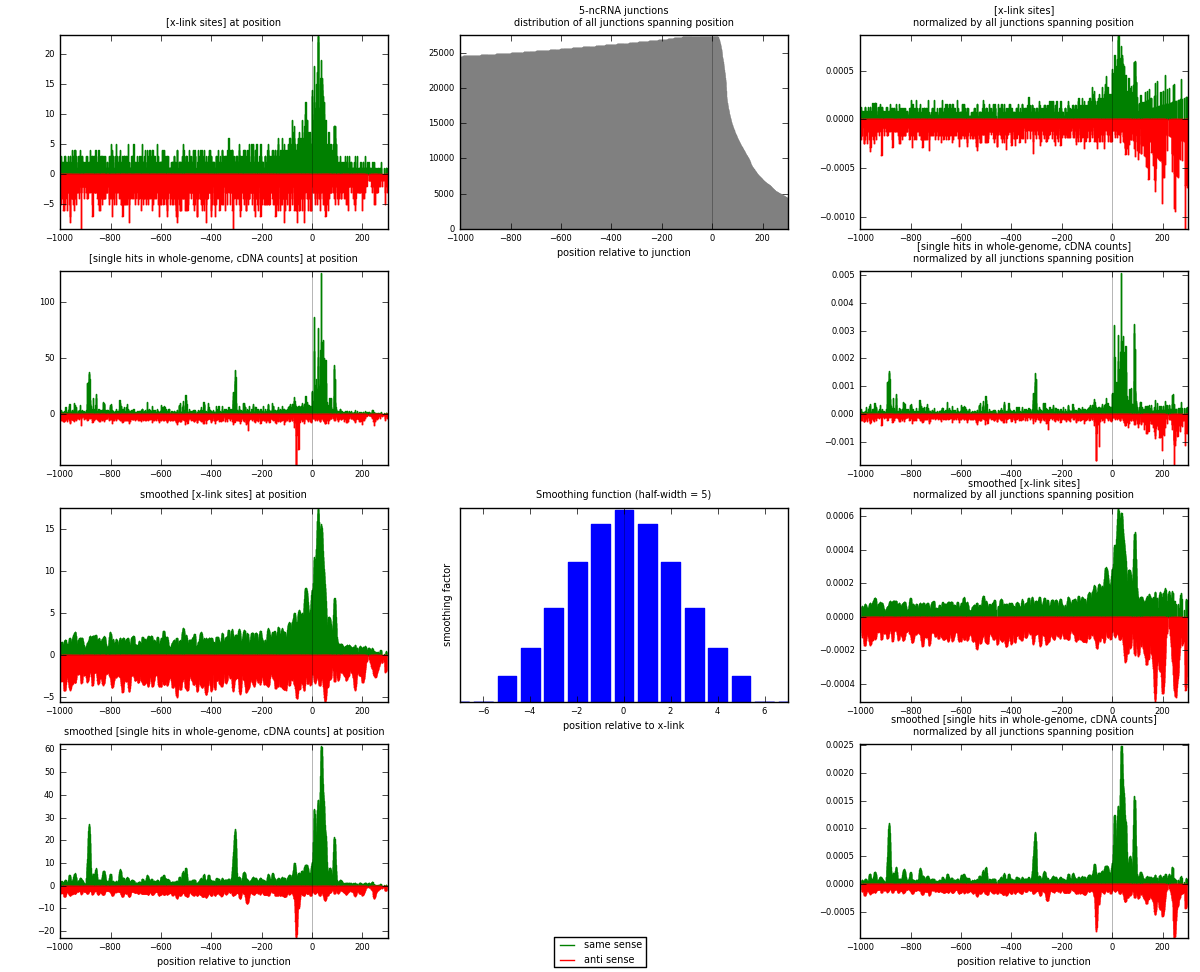 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2957 | 0.68% | 2.17% | 46.21% | 1244 | 1.06712 |
| anti | 3442 | 0.80% | 2.52% | 53.79% | 1583 | 1.24215 |
| both | 6399 | 1.48% | 4.69% | 100.00% | 2771 | 2.30927 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 5500 | 1.16% | 3.54% | 58.69% | 1244 | 1.98484 |
| anti | 3872 | 0.82% | 2.49% | 41.31% | 1583 | 1.39733 |
| both | 9372 | 1.97% | 6.03% | 100.00% | 2771 | 3.38217 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
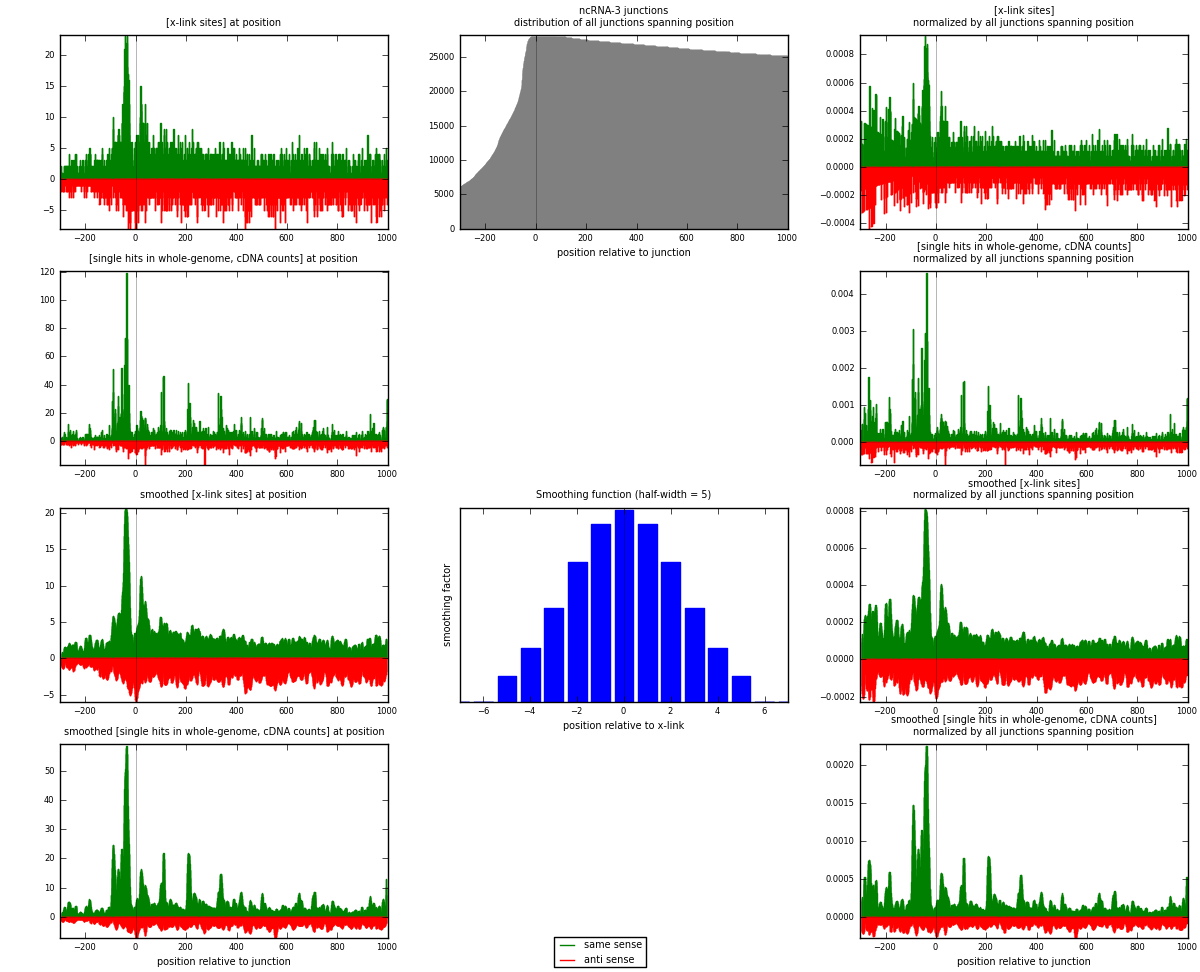 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3585 | 0.83% | 2.63% | 54.36% | 1487 | 1.26634 |
| anti | 3010 | 0.70% | 2.20% | 45.64% | 1386 | 1.06323 |
| both | 6595 | 1.53% | 4.83% | 100.00% | 2831 | 2.32957 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 6326 | 1.33% | 4.07% | 65.71% | 1487 | 2.23455 |
| anti | 3301 | 0.70% | 2.12% | 34.29% | 1386 | 1.16602 |
| both | 9627 | 2.03% | 6.20% | 100.00% | 2831 | 3.40057 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
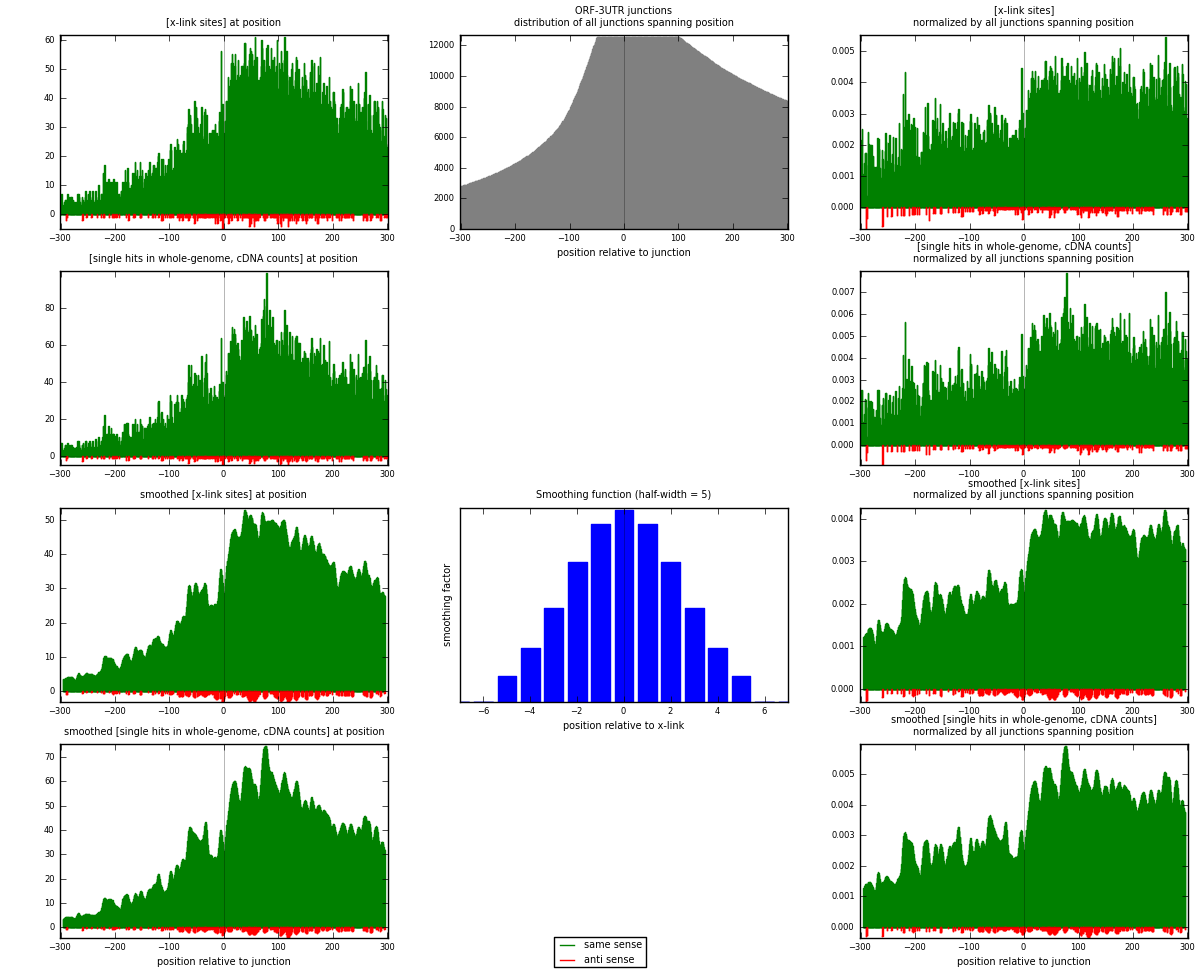
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 16626 | 3.85% | 12.18% | 98.26% | 4109 | 3.91938 |
| anti | 295 | 0.07% | 0.22% | 1.74% | 194 | 0.0695427 |
| both | 16921 | 3.91% | 12.40% | 100.00% | 4242 | 3.98892 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 20263 | 4.27% | 13.04% | 98.48% | 4109 | 4.77676 |
| anti | 313 | 0.07% | 0.20% | 1.52% | 194 | 0.073786 |
| both | 20576 | 4.33% | 13.25% | 100.00% | 4242 | 4.85054 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
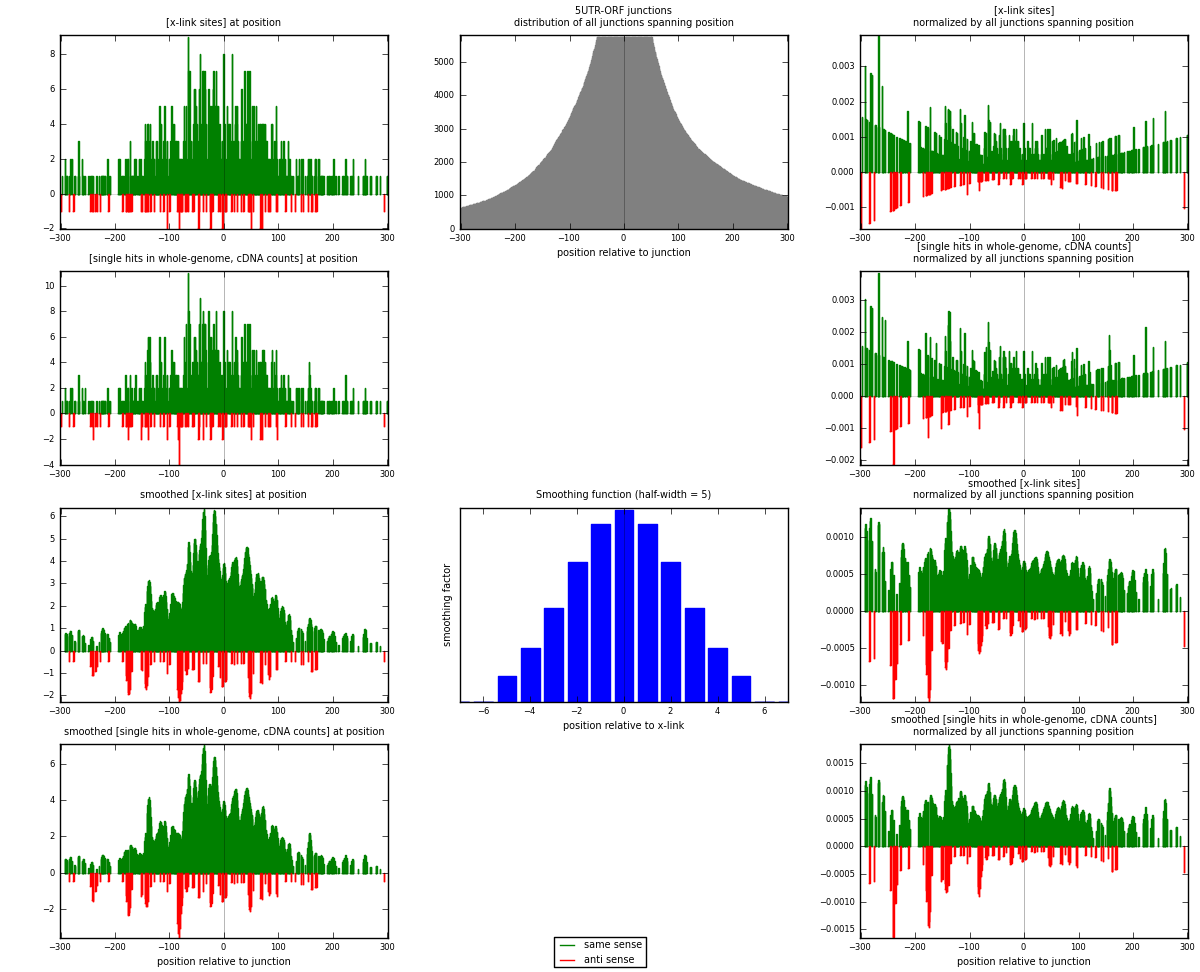
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 877 | 0.20% | 0.64% | 90.13% | 546 | 1.50429 |
| anti | 96 | 0.02% | 0.07% | 9.87% | 41 | 0.164666 |
| both | 973 | 0.23% | 0.71% | 100.00% | 583 | 1.66895 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 929 | 0.20% | 0.60% | 89.85% | 546 | 1.59348 |
| anti | 105 | 0.02% | 0.07% | 10.15% | 41 | 0.180103 |
| both | 1034 | 0.22% | 0.67% | 100.00% | 583 | 1.77358 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
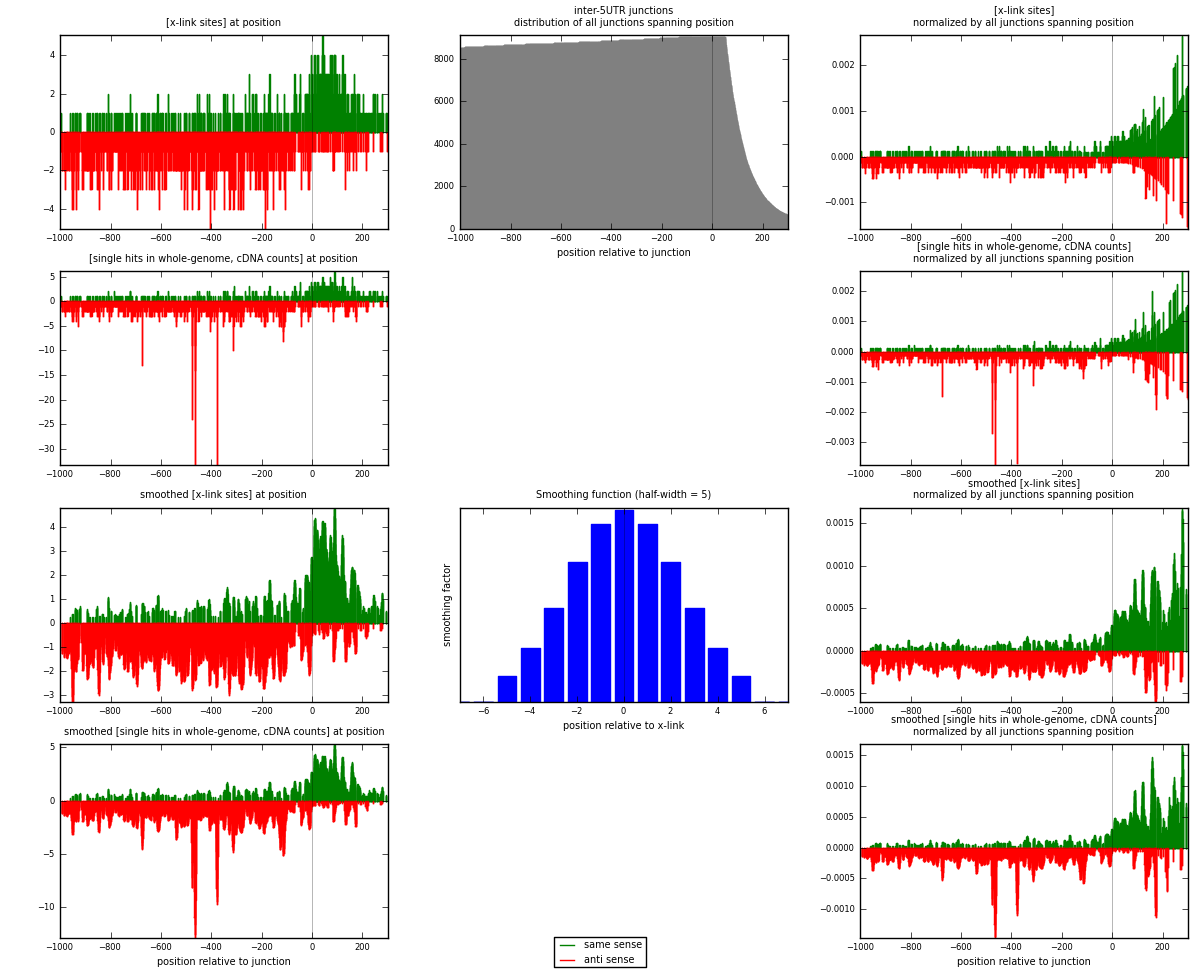
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 641 | 0.15% | 0.47% | 34.89% | 468 | 0.591328 |
| anti | 1196 | 0.28% | 0.88% | 65.11% | 674 | 1.10332 |
| both | 1837 | 0.42% | 1.35% | 100.00% | 1084 | 1.69465 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 663 | 0.14% | 0.43% | 31.23% | 468 | 0.611624 |
| anti | 1460 | 0.31% | 0.94% | 68.77% | 674 | 1.34686 |
| both | 2123 | 0.45% | 1.37% | 100.00% | 1084 | 1.95849 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
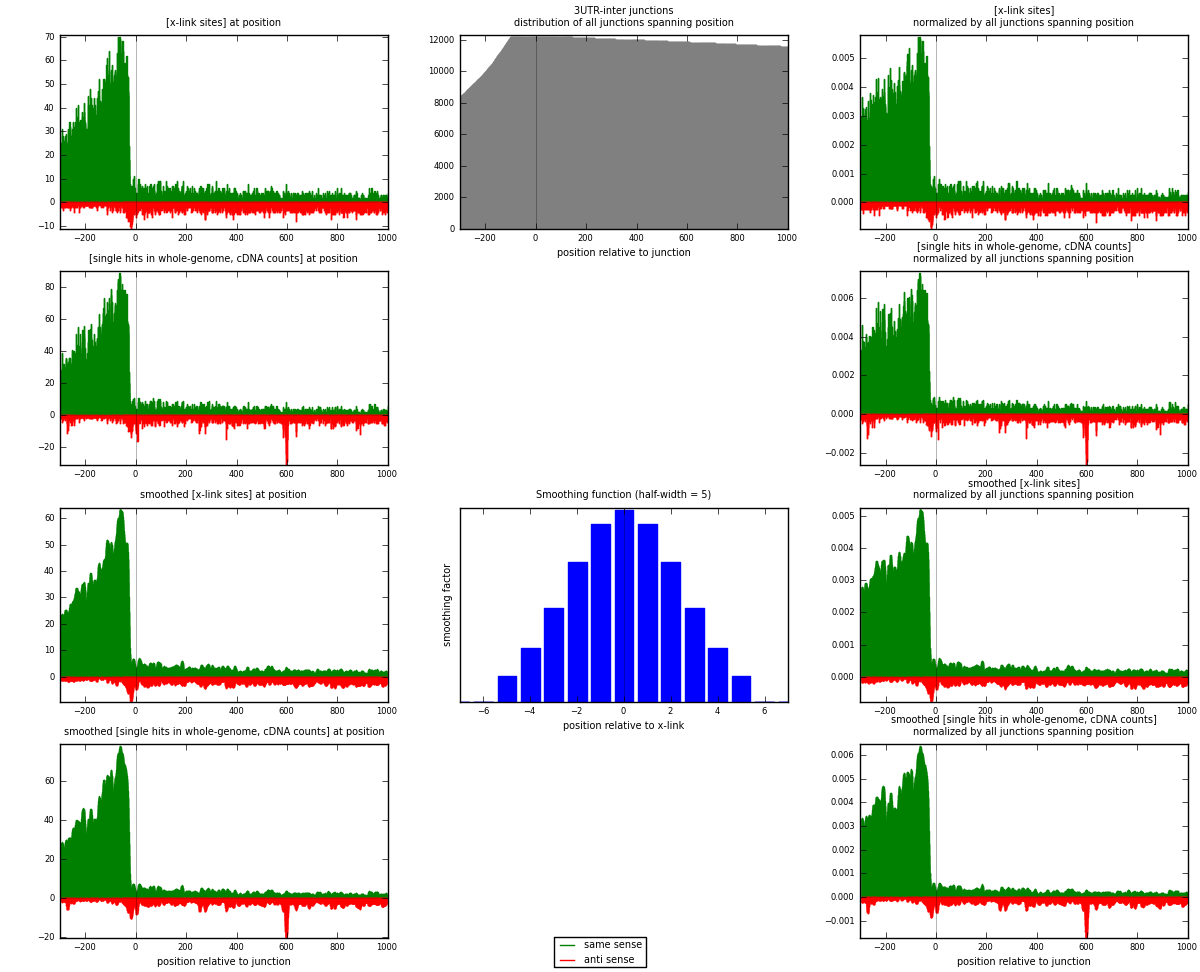
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13452 | 3.11% | 9.85% | 83.34% | 3907 | 3.00469 |
| anti | 2689 | 0.62% | 1.97% | 16.66% | 855 | 0.600625 |
| both | 16141 | 3.73% | 11.82% | 100.00% | 4477 | 3.60532 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 15813 | 3.33% | 10.18% | 82.63% | 3907 | 3.53205 |
| anti | 3323 | 0.70% | 2.14% | 17.37% | 855 | 0.742238 |
| both | 19136 | 4.03% | 12.32% | 100.00% | 4477 | 4.27429 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
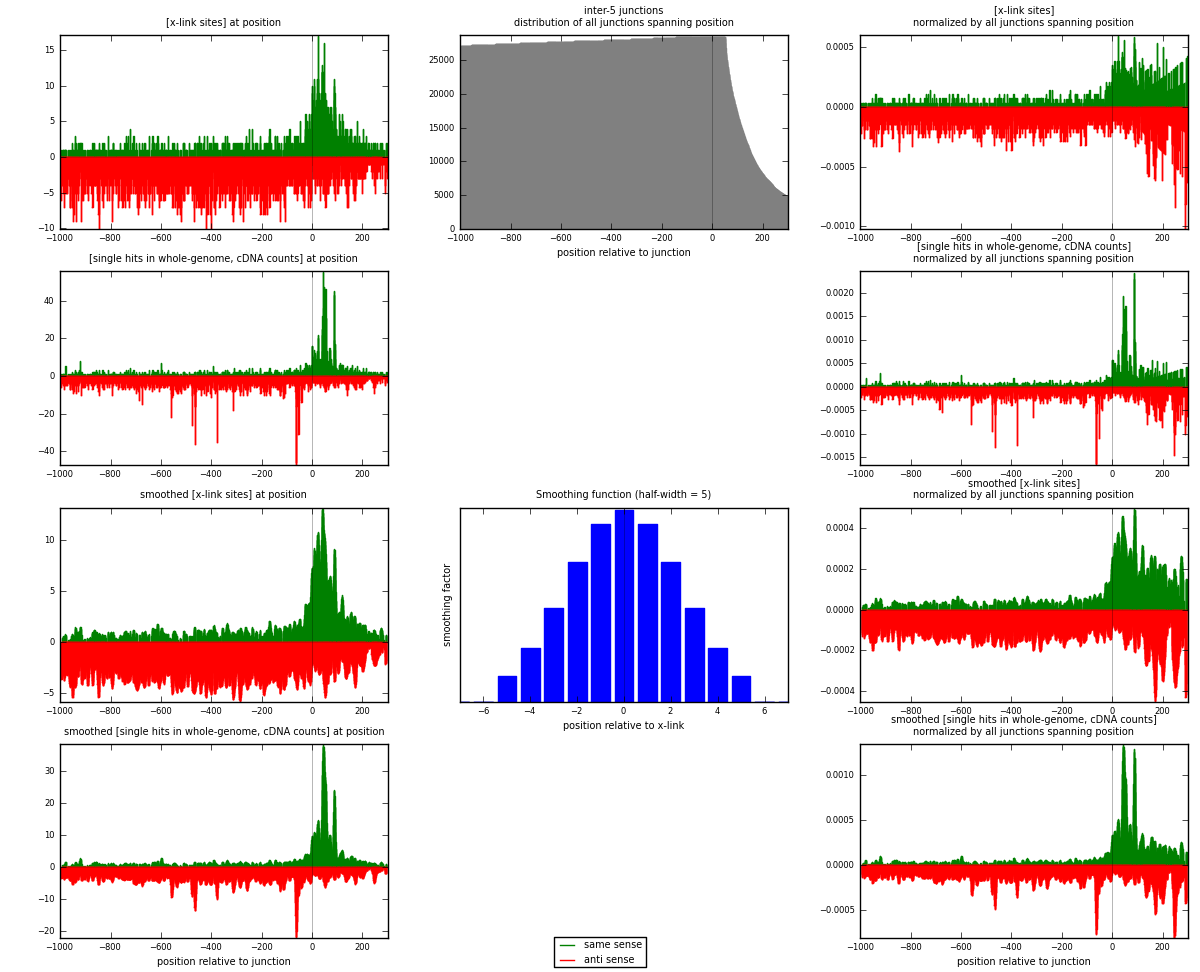
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1671 | 0.39% | 1.22% | 28.55% | 995 | 0.572653 |
| anti | 4181 | 0.97% | 3.06% | 71.45% | 2032 | 1.43283 |
| both | 5852 | 1.35% | 4.29% | 100.00% | 2918 | 2.00548 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2304 | 0.49% | 1.48% | 31.85% | 995 | 0.789582 |
| anti | 4930 | 1.04% | 3.17% | 68.15% | 2032 | 1.68951 |
| both | 7234 | 1.52% | 4.66% | 100.00% | 2918 | 2.4791 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
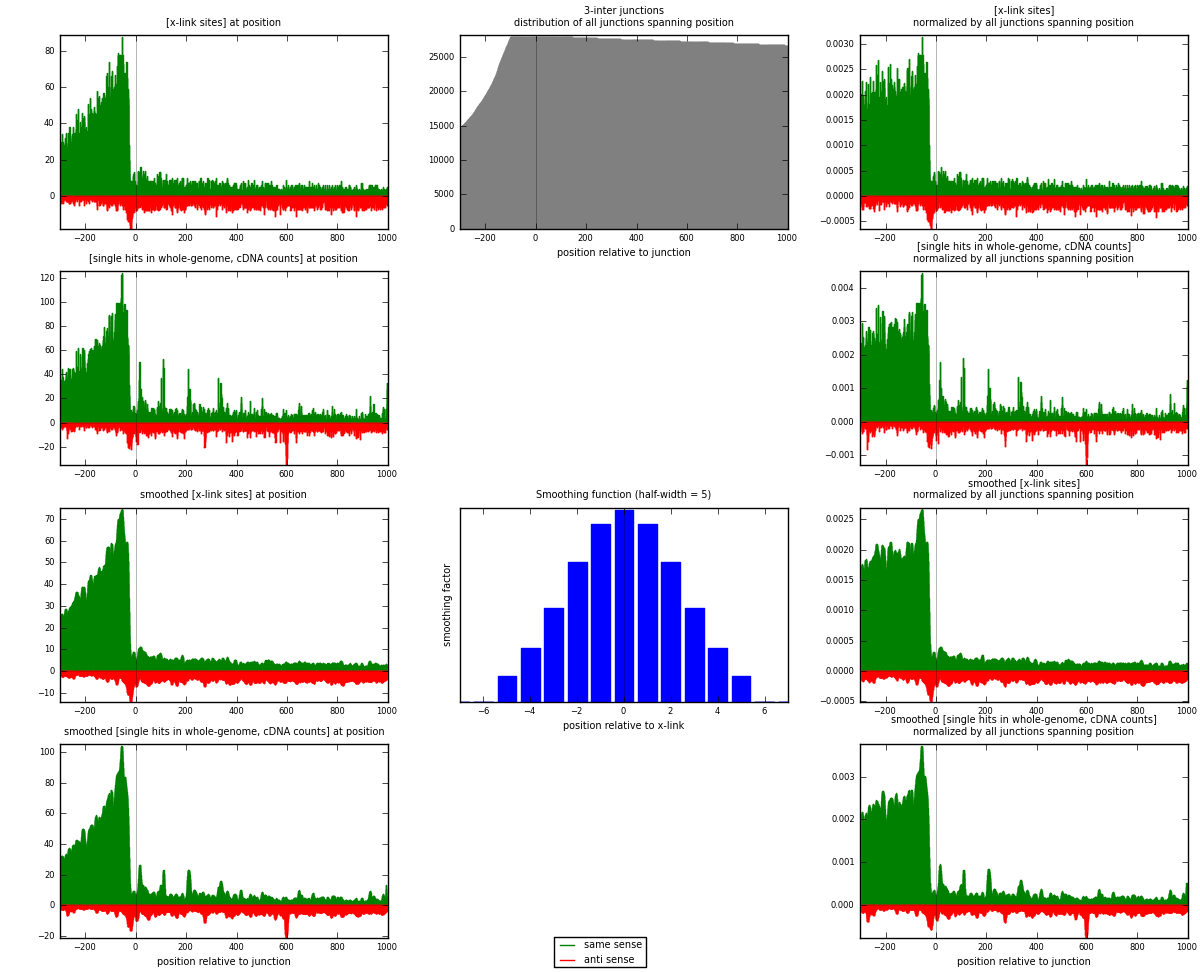
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 16413 | 3.80% | 12.02% | 75.83% | 4815 | 2.60938 |
| anti | 5232 | 1.21% | 3.83% | 24.17% | 1822 | 0.831797 |
| both | 21645 | 5.01% | 15.86% | 100.00% | 6290 | 3.44118 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 21267 | 4.48% | 13.69% | 77.37% | 4815 | 3.38108 |
| anti | 6221 | 1.31% | 4.00% | 22.63% | 1822 | 0.98903 |
| both | 27488 | 5.79% | 17.70% | 100.00% | 6290 | 4.37011 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
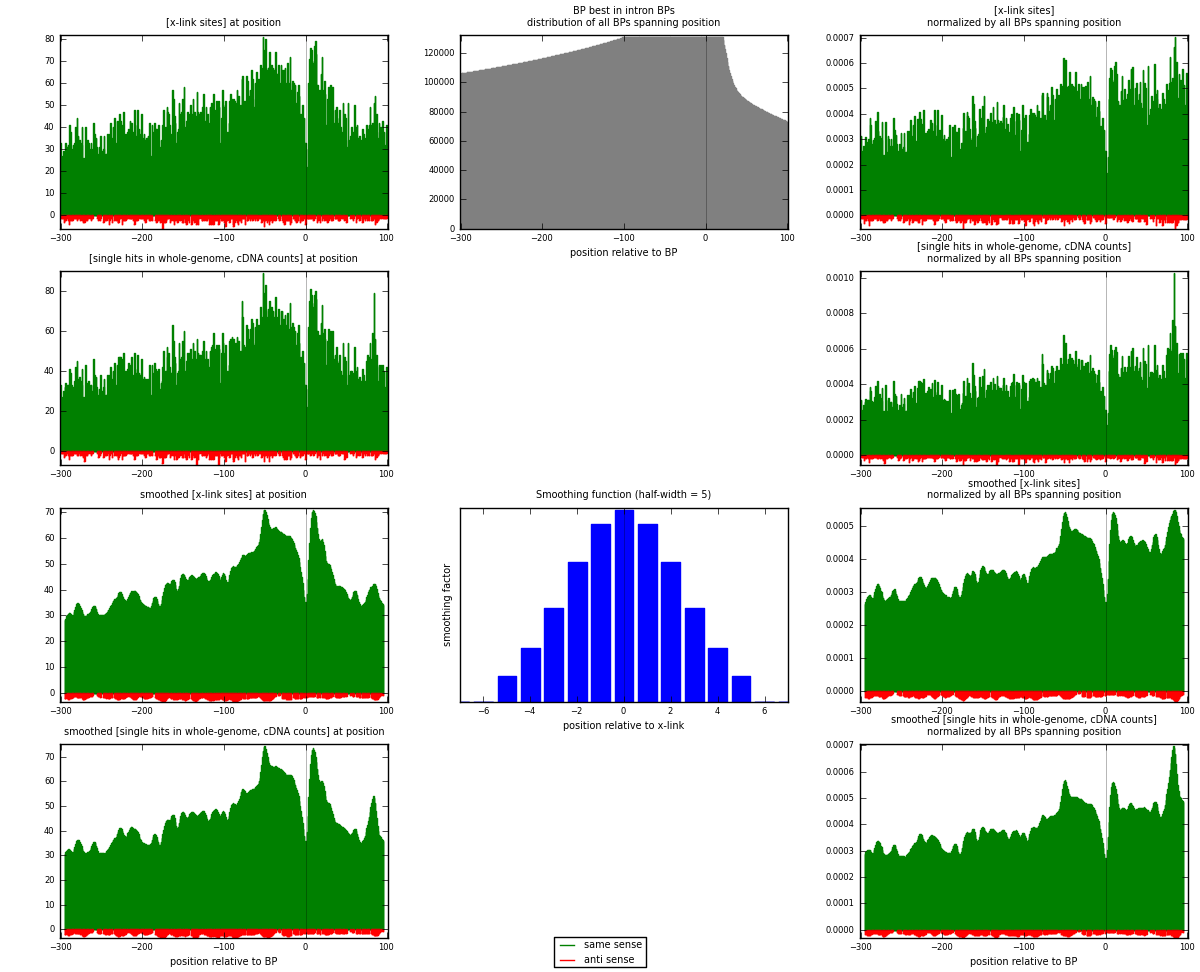
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 17516 | 4.05% | 12.83% | 96.74% | 12928 | 1.30971 |
| anti | 590 | 0.14% | 0.43% | 3.26% | 474 | 0.0441154 |
| both | 18106 | 4.19% | 13.26% | 100.00% | 13374 | 1.35382 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 18236 | 3.84% | 11.74% | 96.71% | 12928 | 1.36354 |
| anti | 621 | 0.13% | 0.40% | 3.29% | 474 | 0.0464334 |
| both | 18857 | 3.97% | 12.14% | 100.00% | 13374 | 1.40997 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.