RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
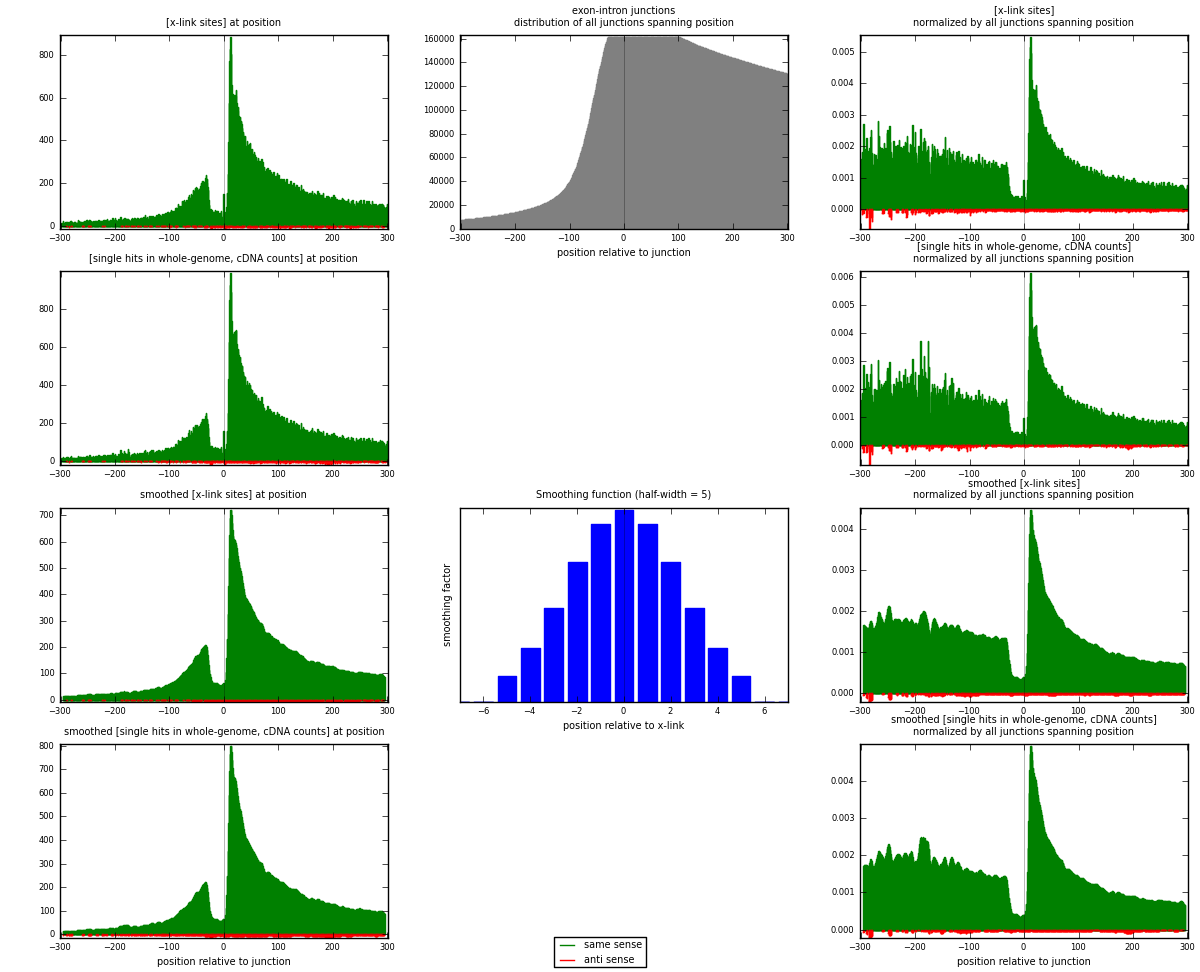 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 79114 | 10.03% | 33.19% | 97.91% | 39030 | 1.98201 |
| anti | 1687 | 0.21% | 0.71% | 2.09% | 1117 | 0.0422638 |
| both | 80801 | 10.24% | 33.89% | 100.00% | 39916 | 2.02428 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 83785 | 9.59% | 30.64% | 97.64% | 39030 | 2.09903 |
| anti | 2024 | 0.23% | 0.74% | 2.36% | 1117 | 0.0507065 |
| both | 85809 | 9.82% | 31.38% | 100.00% | 39916 | 2.14974 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 56911 | 7.21% | 23.87% | 96.79% | 31720 | 1.73615 |
| anti | 1886 | 0.24% | 0.79% | 3.21% | 1312 | 0.0575351 |
| both | 58797 | 7.45% | 24.66% | 100.00% | 32780 | 1.79369 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 60936 | 6.97% | 22.29% | 96.79% | 31720 | 1.85894 |
| anti | 2024 | 0.23% | 0.74% | 3.21% | 1312 | 0.061745 |
| both | 62960 | 7.20% | 23.03% | 100.00% | 32780 | 1.92068 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
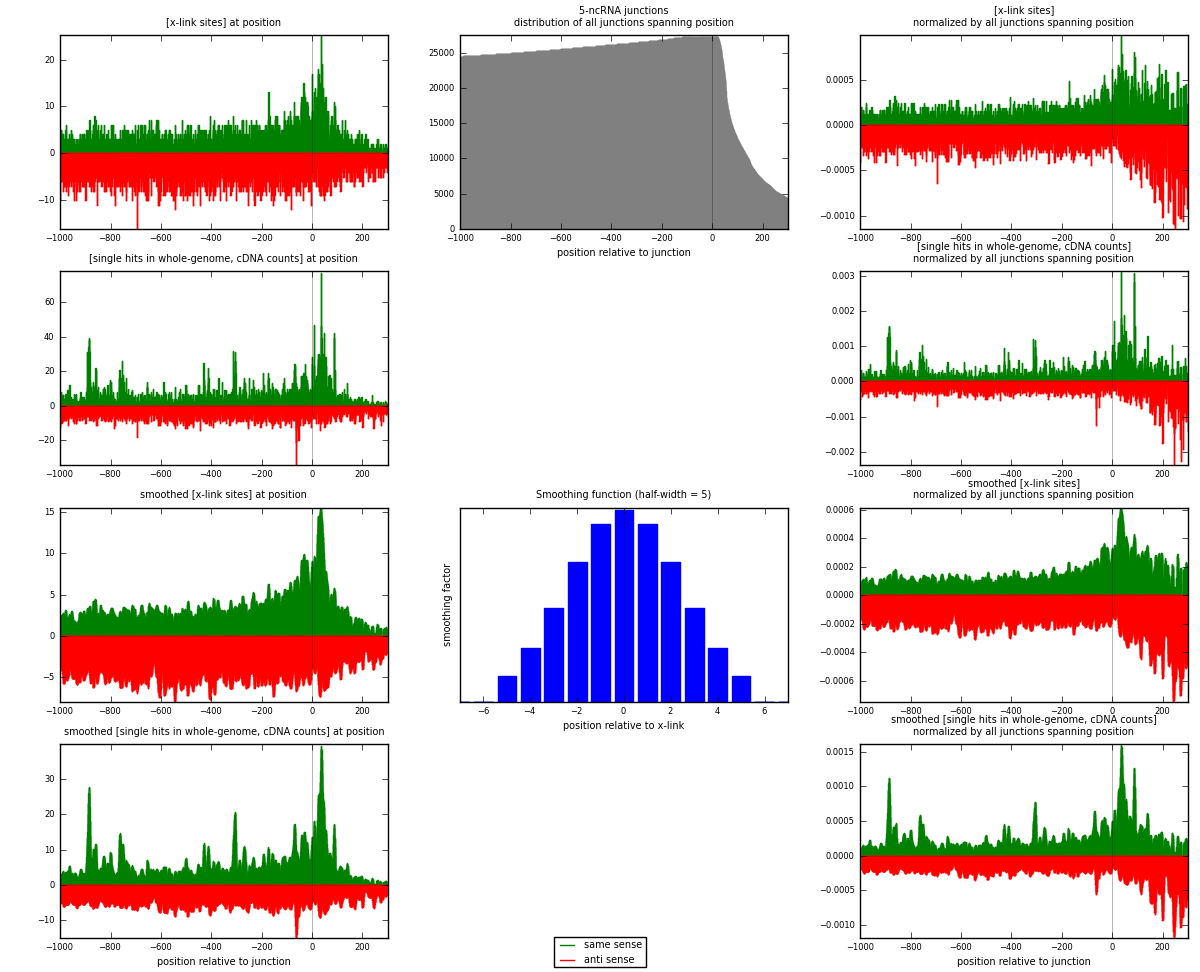 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4487 | 0.57% | 1.88% | 41.45% | 1777 | 1.14875 |
| anti | 6339 | 0.80% | 2.66% | 58.55% | 2237 | 1.62289 |
| both | 10826 | 1.37% | 4.54% | 100.00% | 3906 | 2.77163 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 7426 | 0.85% | 2.72% | 50.99% | 1777 | 1.90118 |
| anti | 7137 | 0.82% | 2.61% | 49.01% | 2237 | 1.82719 |
| both | 14563 | 1.67% | 5.33% | 100.00% | 3906 | 3.72837 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
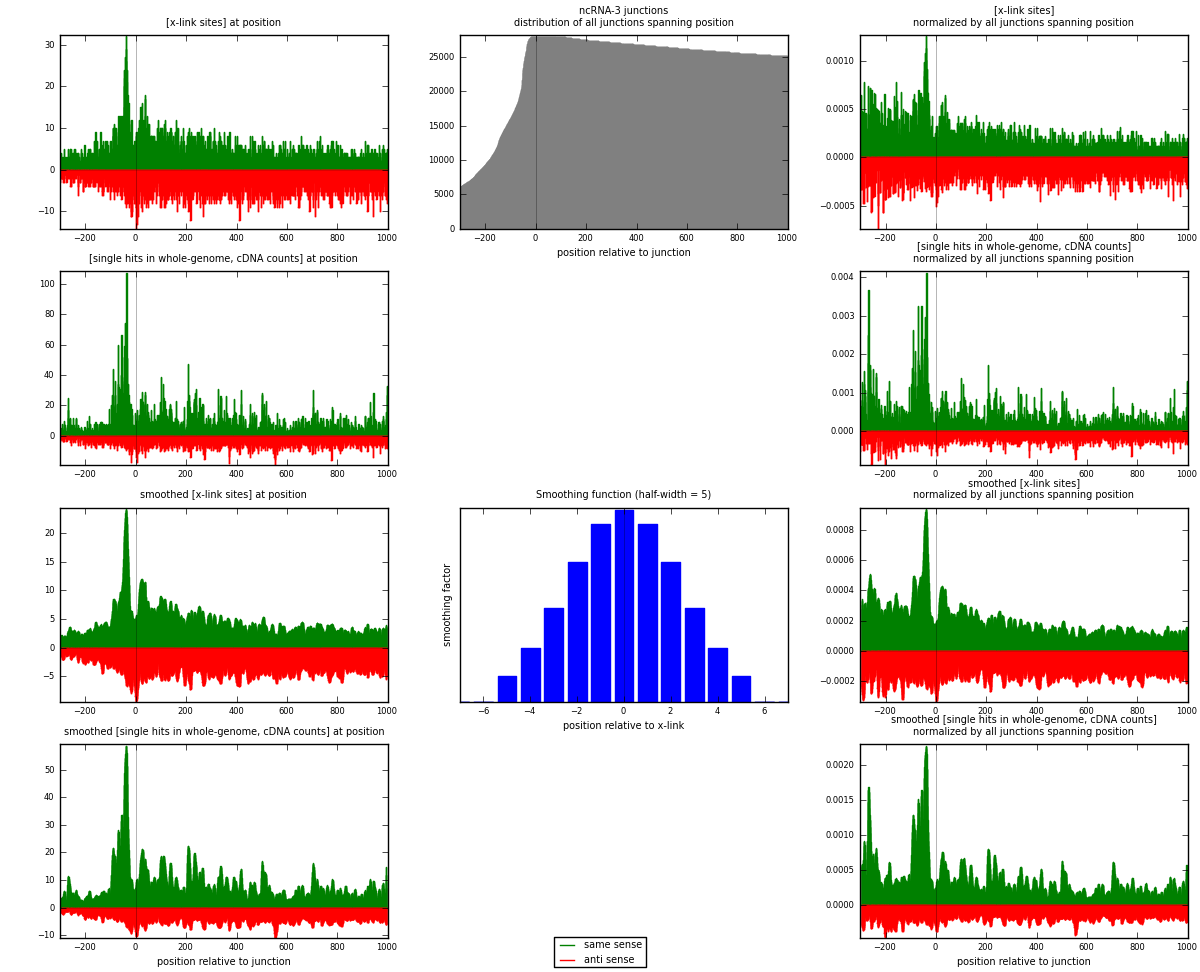 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5845 | 0.74% | 2.45% | 51.96% | 2075 | 1.47452 |
| anti | 5404 | 0.69% | 2.27% | 48.04% | 1978 | 1.36327 |
| both | 11249 | 1.43% | 4.72% | 100.00% | 3964 | 2.83779 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 10715 | 1.23% | 3.92% | 64.31% | 2075 | 2.70308 |
| anti | 5946 | 0.68% | 2.17% | 35.69% | 1978 | 1.5 |
| both | 16661 | 1.91% | 6.09% | 100.00% | 3964 | 4.20308 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
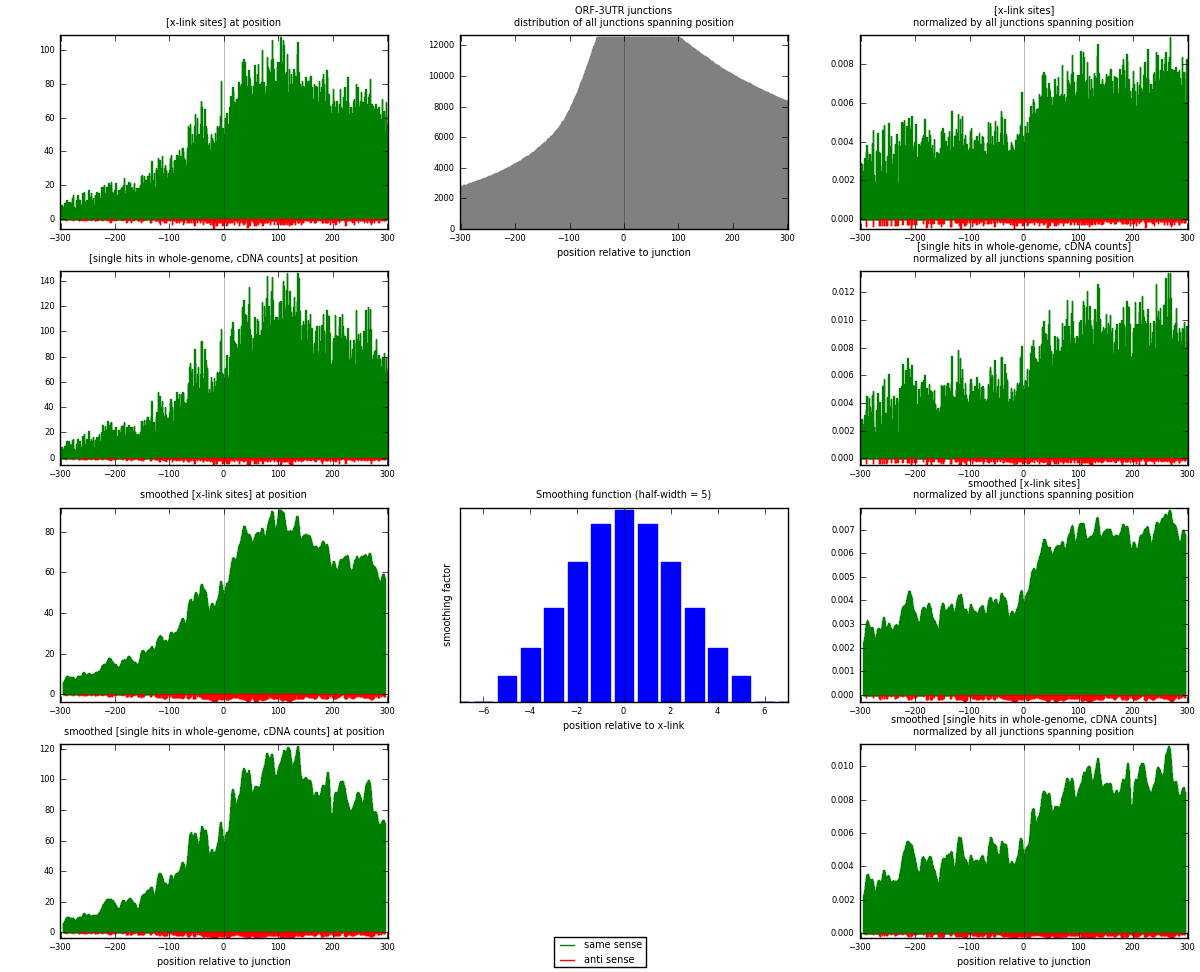 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 29193 | 3.70% | 12.25% | 98.23% | 4976 | 5.65975 |
| anti | 525 | 0.07% | 0.22% | 1.77% | 290 | 0.101784 |
| both | 29718 | 3.77% | 12.47% | 100.00% | 5158 | 5.76154 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 37754 | 4.32% | 13.81% | 98.54% | 4976 | 7.3195 |
| anti | 558 | 0.06% | 0.20% | 1.46% | 290 | 0.108181 |
| both | 38312 | 4.38% | 14.01% | 100.00% | 5158 | 7.42769 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
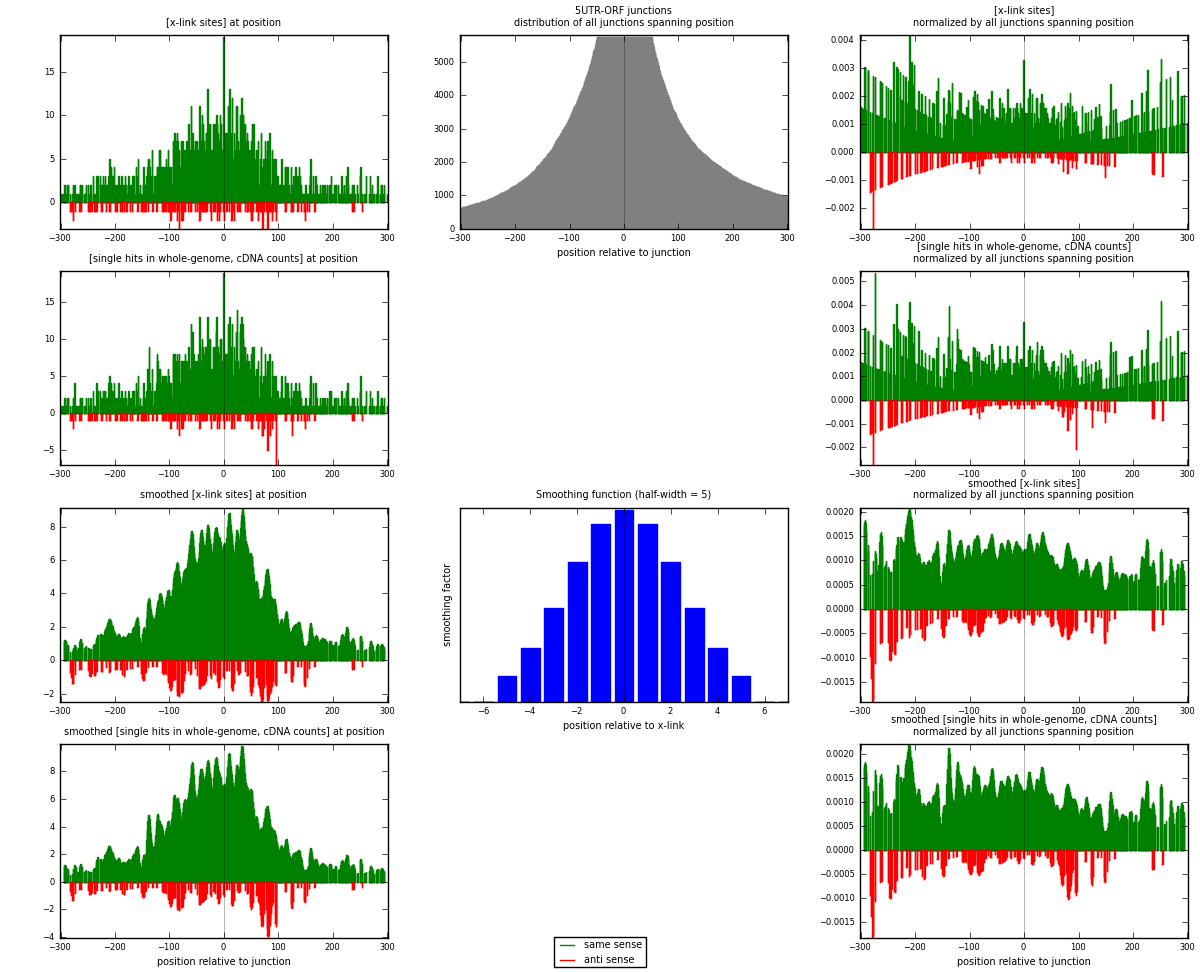 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1718 | 0.22% | 0.72% | 91.24% | 850 | 1.89625 |
| anti | 165 | 0.02% | 0.07% | 8.76% | 67 | 0.182119 |
| both | 1883 | 0.24% | 0.79% | 100.00% | 906 | 2.07837 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1828 | 0.21% | 0.67% | 90.99% | 850 | 2.01766 |
| anti | 181 | 0.02% | 0.07% | 9.01% | 67 | 0.199779 |
| both | 2009 | 0.23% | 0.73% | 100.00% | 906 | 2.21744 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
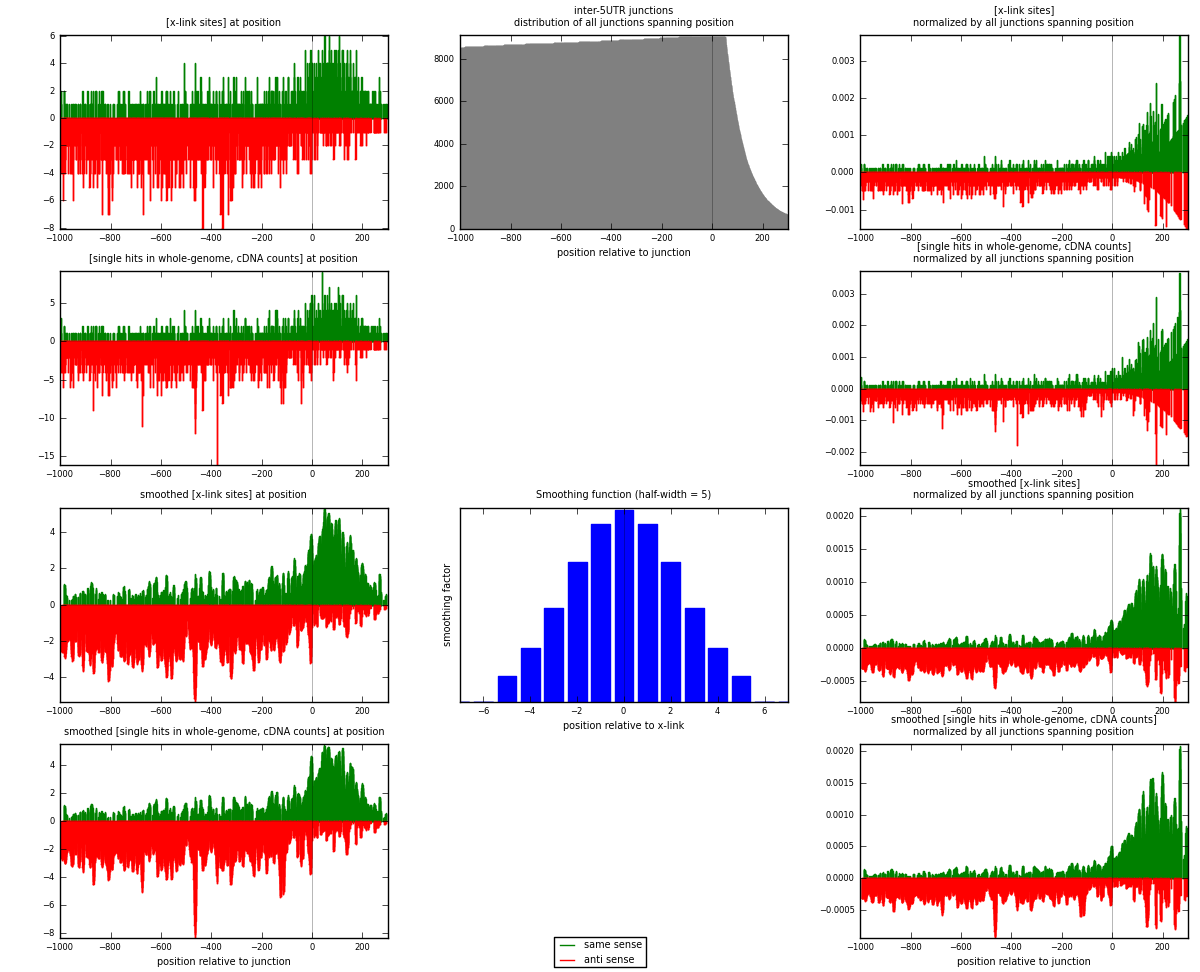 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1082 | 0.14% | 0.45% | 33.05% | 654 | 0.68179 |
| anti | 2192 | 0.28% | 0.92% | 66.95% | 1042 | 1.38122 |
| both | 3274 | 0.42% | 1.37% | 100.00% | 1587 | 2.06301 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1145 | 0.13% | 0.42% | 31.88% | 654 | 0.721487 |
| anti | 2447 | 0.28% | 0.89% | 68.12% | 1042 | 1.5419 |
| both | 3592 | 0.41% | 1.31% | 100.00% | 1587 | 2.26339 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
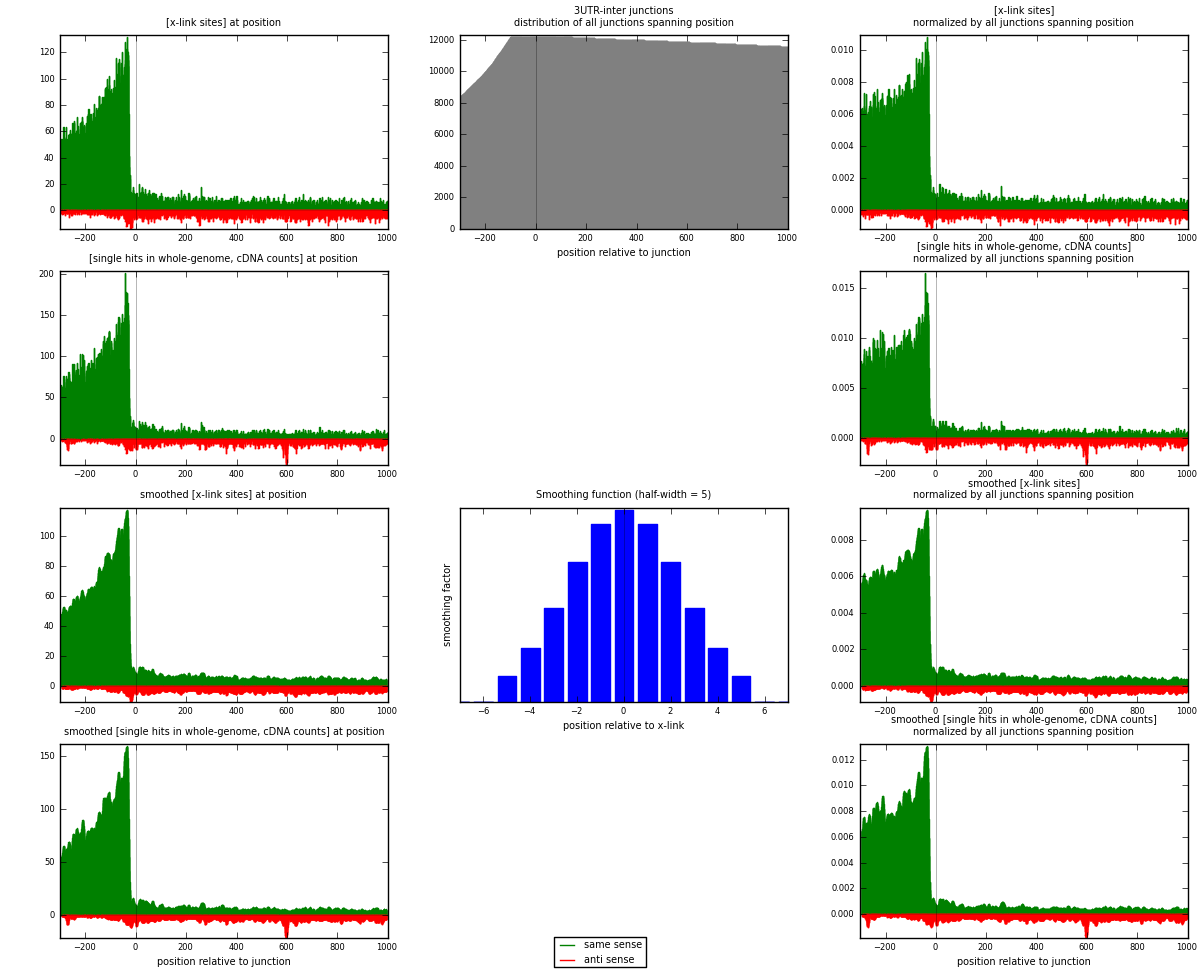 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 25828 | 3.27% | 10.83% | 84.89% | 4861 | 4.64532 |
| anti | 4598 | 0.58% | 1.93% | 15.11% | 1104 | 0.826978 |
| both | 30426 | 3.86% | 12.76% | 100.00% | 5560 | 5.4723 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 31784 | 3.64% | 11.62% | 84.69% | 4861 | 5.71655 |
| anti | 5747 | 0.66% | 2.10% | 15.31% | 1104 | 1.03363 |
| both | 37531 | 4.29% | 13.73% | 100.00% | 5560 | 6.75018 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
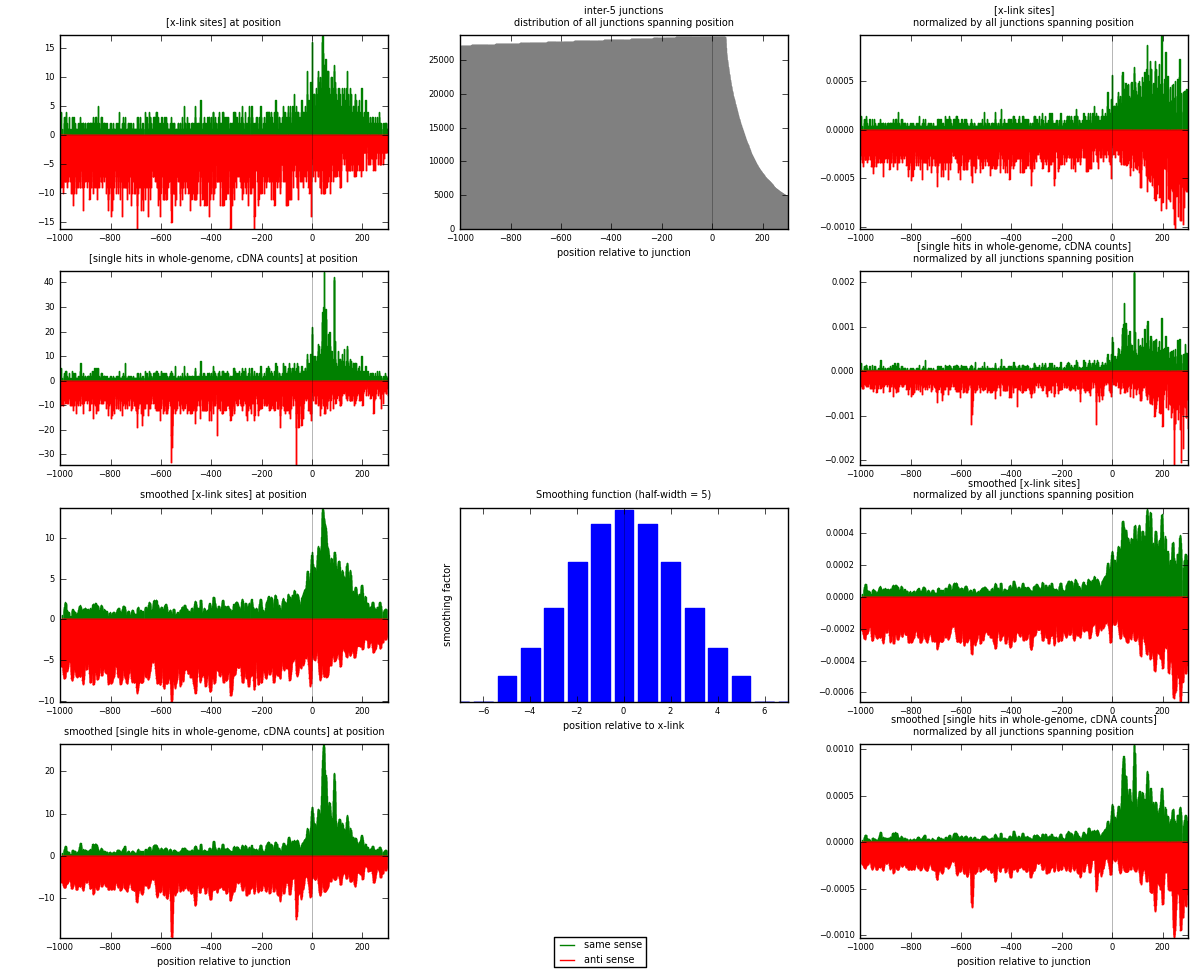 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2642 | 0.33% | 1.11% | 25.57% | 1403 | 0.63236 |
| anti | 7689 | 0.97% | 3.23% | 74.43% | 2972 | 1.84035 |
| both | 10331 | 1.31% | 4.33% | 100.00% | 4178 | 2.47271 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3256 | 0.37% | 1.19% | 26.96% | 1403 | 0.77932 |
| anti | 8822 | 1.01% | 3.23% | 73.04% | 2972 | 2.11154 |
| both | 12078 | 1.38% | 4.42% | 100.00% | 4178 | 2.89086 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
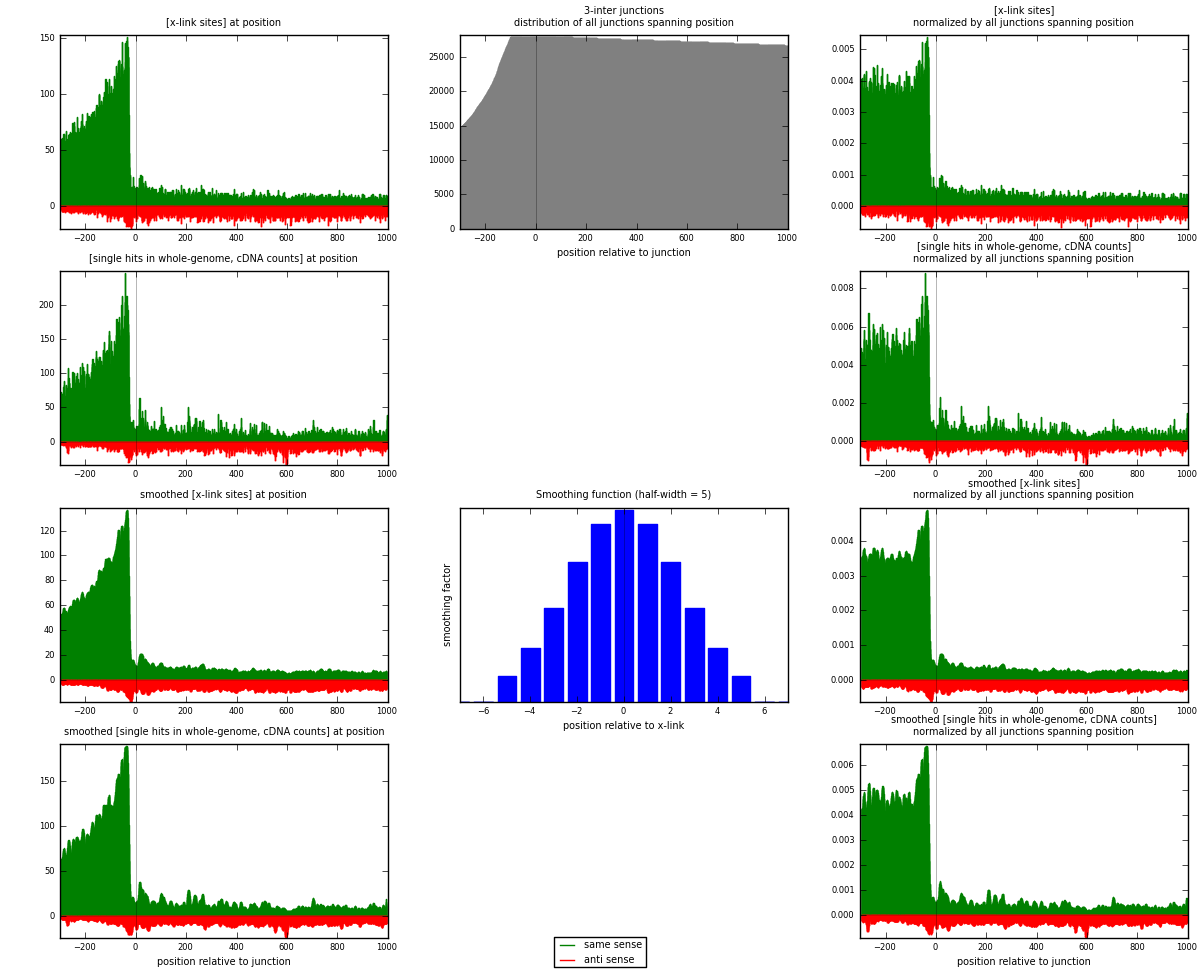 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 30955 | 3.92% | 12.98% | 77.25% | 6170 | 3.82727 |
| anti | 9117 | 1.16% | 3.82% | 22.75% | 2441 | 1.12723 |
| both | 40072 | 5.08% | 16.81% | 100.00% | 8088 | 4.9545 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 41892 | 4.79% | 15.32% | 79.20% | 6170 | 5.17953 |
| anti | 10999 | 1.26% | 4.02% | 20.80% | 2441 | 1.35992 |
| both | 52891 | 6.05% | 19.34% | 100.00% | 8088 | 6.53944 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
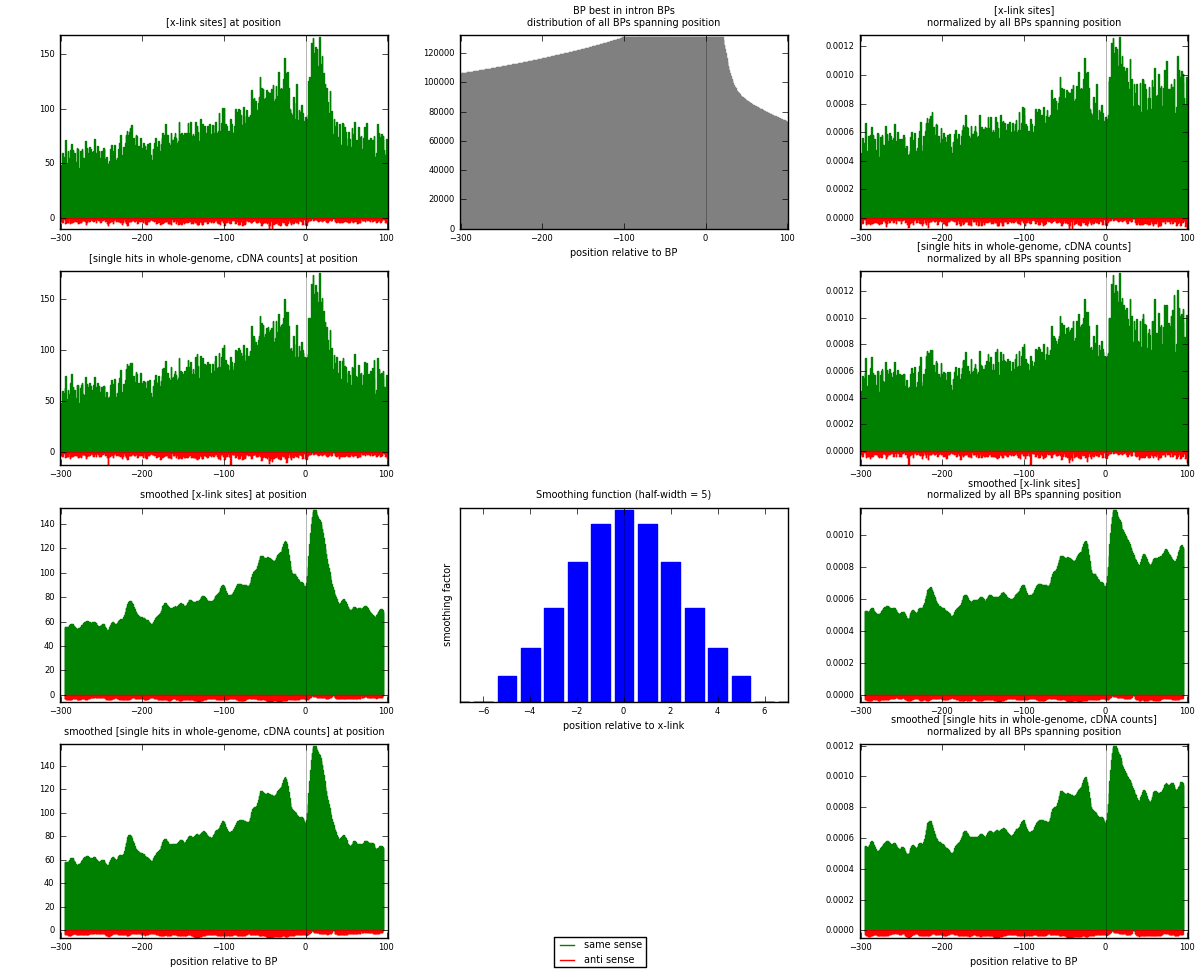 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 32341 | 4.10% | 13.57% | 96.59% | 20723 | 1.50605 |
| anti | 1141 | 0.14% | 0.48% | 3.41% | 881 | 0.053134 |
| both | 33482 | 4.24% | 14.04% | 100.00% | 21474 | 1.55919 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 33548 | 3.84% | 12.27% | 96.54% | 20723 | 1.56226 |
| anti | 1204 | 0.14% | 0.44% | 3.46% | 881 | 0.0560678 |
| both | 34752 | 3.98% | 12.71% | 100.00% | 21474 | 1.61833 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.