RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
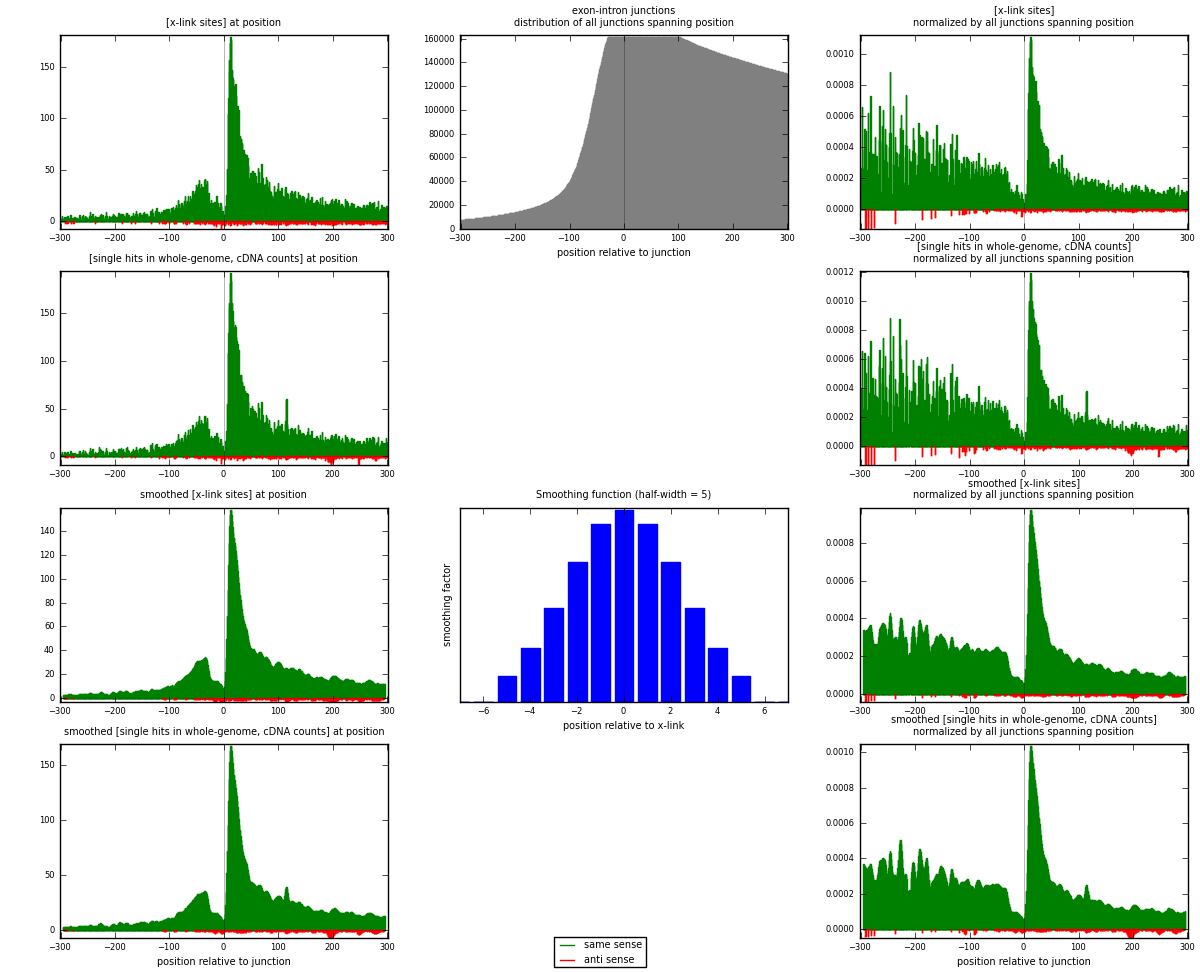 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12258 | 9.19% | 28.45% | 97.51% | 9878 | 1.21643 |
| anti | 313 | 0.23% | 0.73% | 2.49% | 225 | 0.0310608 |
| both | 12571 | 9.43% | 29.17% | 100.00% | 10077 | 1.24749 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 12966 | 8.79% | 26.33% | 97.17% | 9878 | 1.28669 |
| anti | 378 | 0.26% | 0.77% | 2.83% | 225 | 0.0375112 |
| both | 13344 | 9.05% | 27.09% | 100.00% | 10077 | 1.3242 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
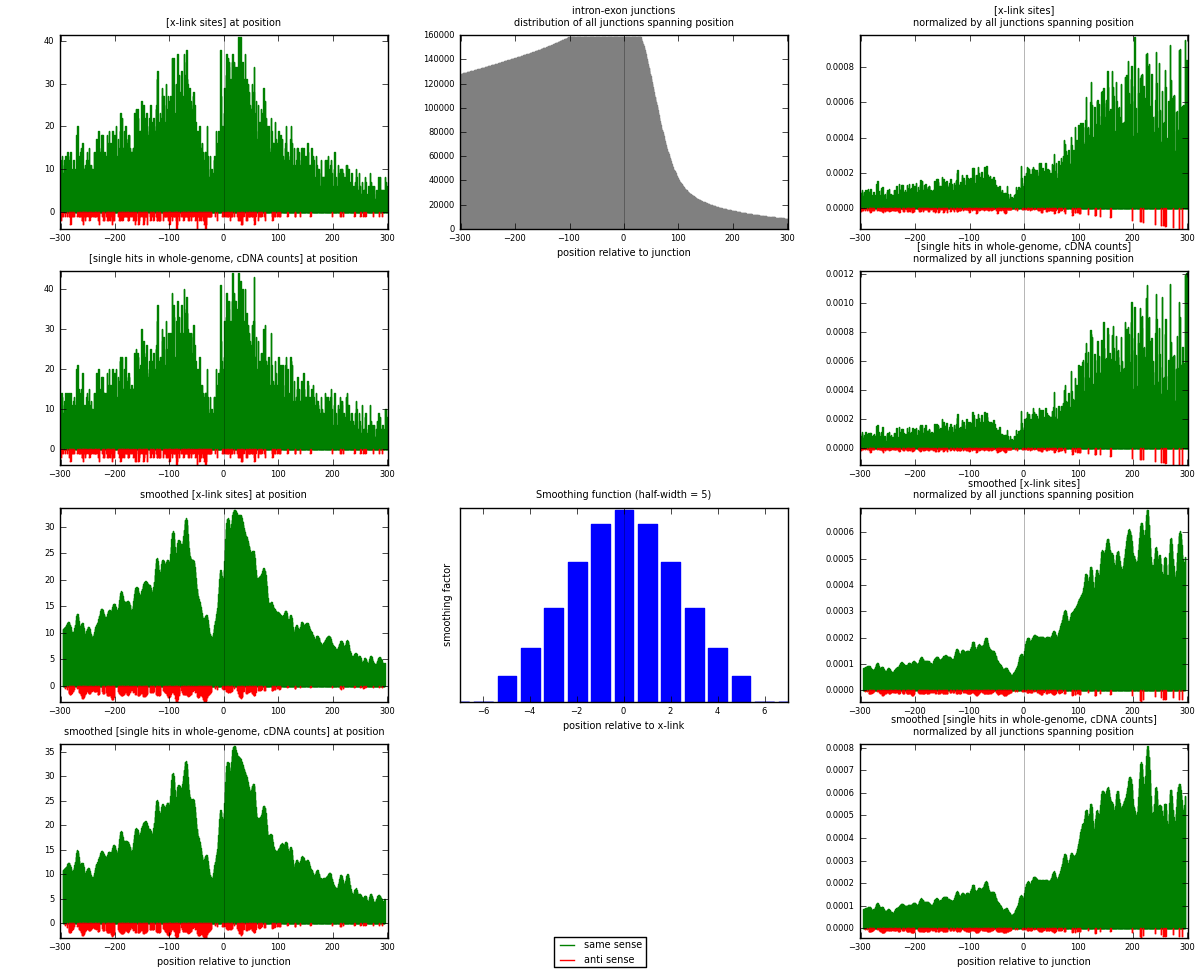 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9162 | 6.87% | 21.26% | 96.54% | 7236 | 1.2229 |
| anti | 328 | 0.25% | 0.76% | 3.46% | 267 | 0.04378 |
| both | 9490 | 7.12% | 22.02% | 100.00% | 7492 | 1.26668 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9750 | 6.61% | 19.80% | 96.67% | 7236 | 1.30139 |
| anti | 336 | 0.23% | 0.68% | 3.33% | 267 | 0.0448478 |
| both | 10086 | 6.84% | 20.48% | 100.00% | 7492 | 1.34624 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1162 | 0.87% | 2.70% | 49.45% | 597 | 0.922222 |
| anti | 1188 | 0.89% | 2.76% | 50.55% | 675 | 0.942857 |
| both | 2350 | 1.76% | 5.45% | 100.00% | 1260 | 1.86508 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2354 | 1.60% | 4.78% | 62.34% | 597 | 1.86825 |
| anti | 1422 | 0.96% | 2.89% | 37.66% | 675 | 1.12857 |
| both | 3776 | 2.56% | 7.67% | 100.00% | 1260 | 2.99683 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
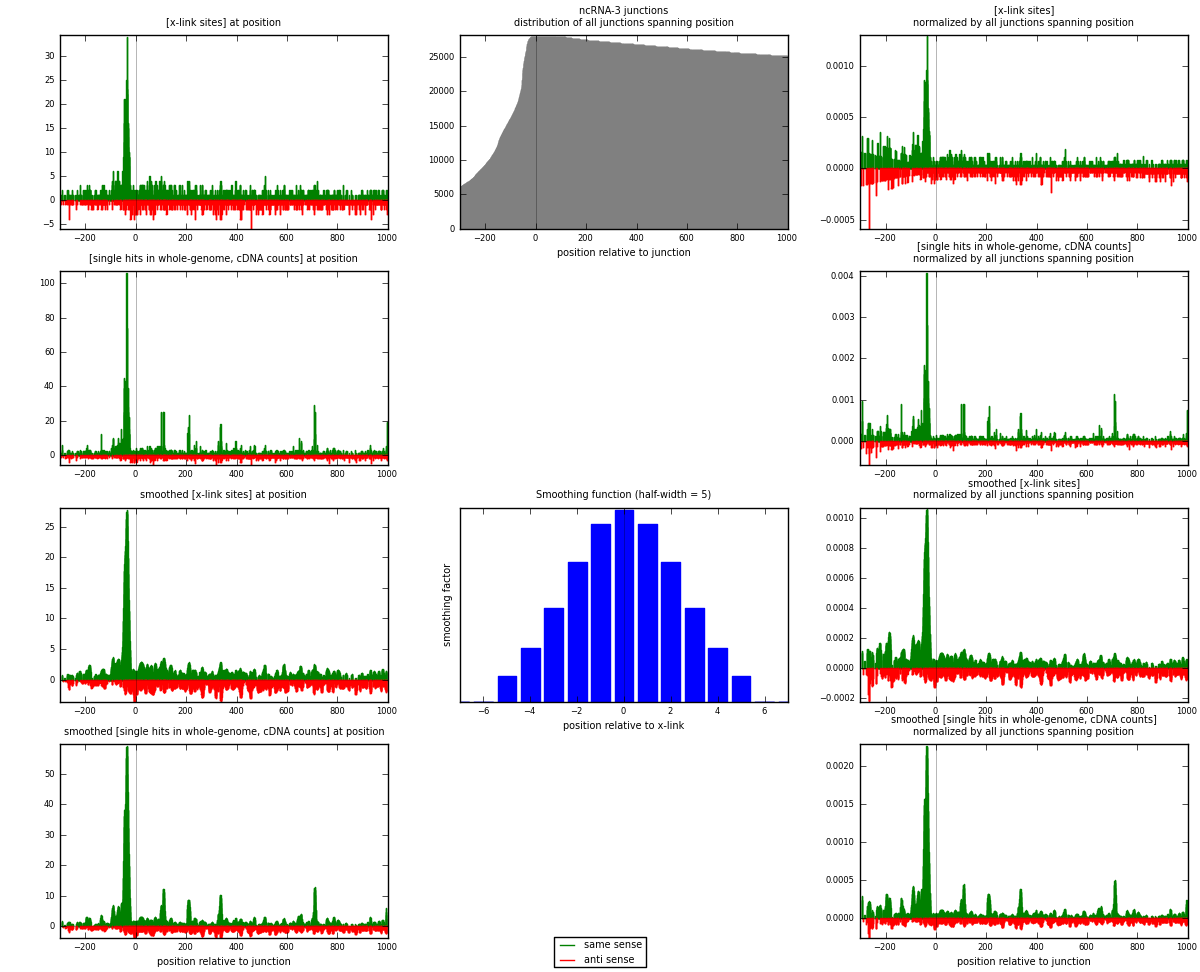 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1582 | 1.19% | 3.67% | 61.97% | 744 | 1.18413 |
| anti | 971 | 0.73% | 2.25% | 38.03% | 604 | 0.726796 |
| both | 2553 | 1.91% | 5.92% | 100.00% | 1336 | 1.91093 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2611 | 1.77% | 5.30% | 71.34% | 744 | 1.95434 |
| anti | 1049 | 0.71% | 2.13% | 28.66% | 604 | 0.78518 |
| both | 3660 | 2.48% | 7.43% | 100.00% | 1336 | 2.73952 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
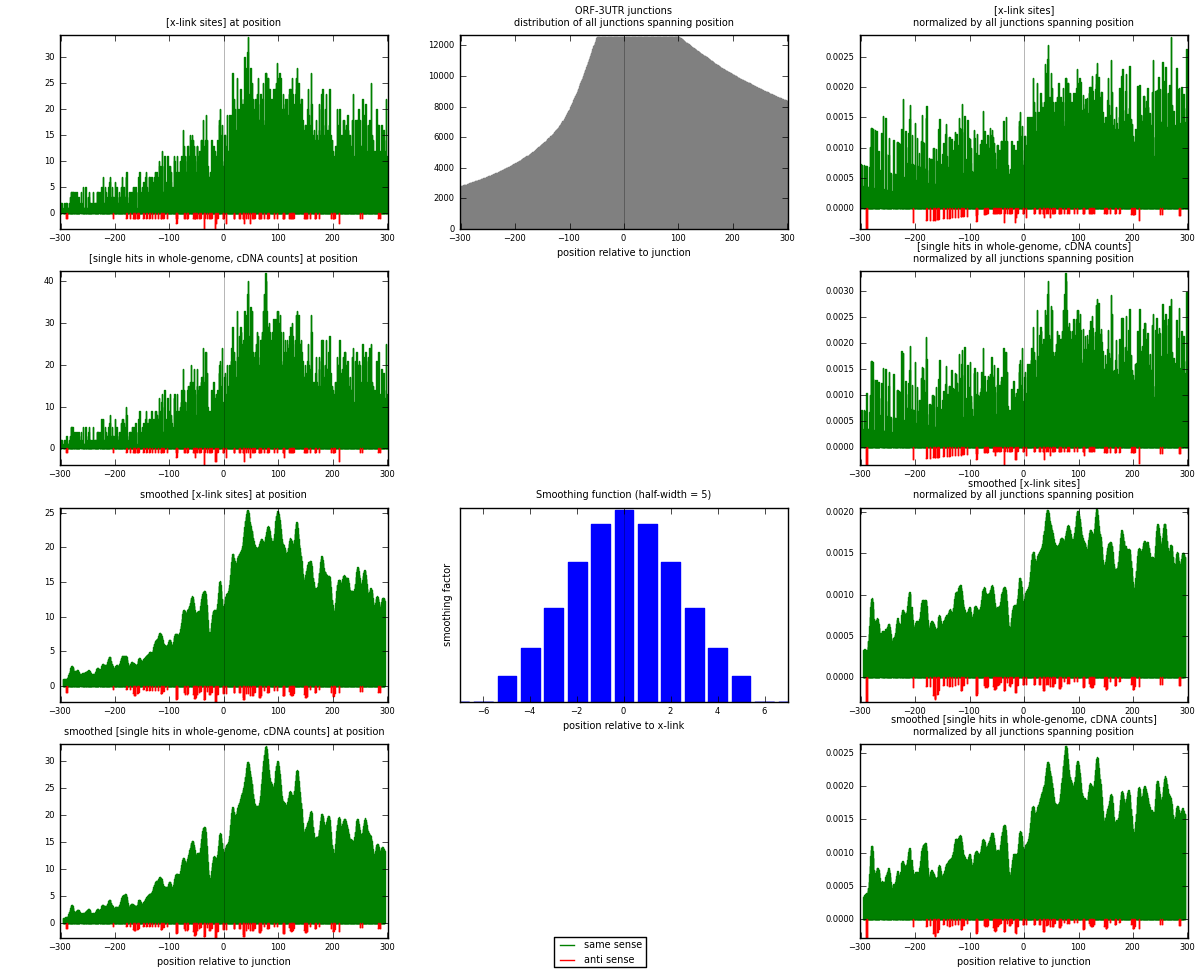 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6920 | 5.19% | 16.06% | 98.53% | 2552 | 2.65236 |
| anti | 103 | 0.08% | 0.24% | 1.47% | 69 | 0.0394787 |
| both | 7023 | 5.27% | 16.30% | 100.00% | 2609 | 2.69184 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 8006 | 5.43% | 16.26% | 98.66% | 2552 | 3.06861 |
| anti | 109 | 0.07% | 0.22% | 1.34% | 69 | 0.0417785 |
| both | 8115 | 5.50% | 16.48% | 100.00% | 2609 | 3.11039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
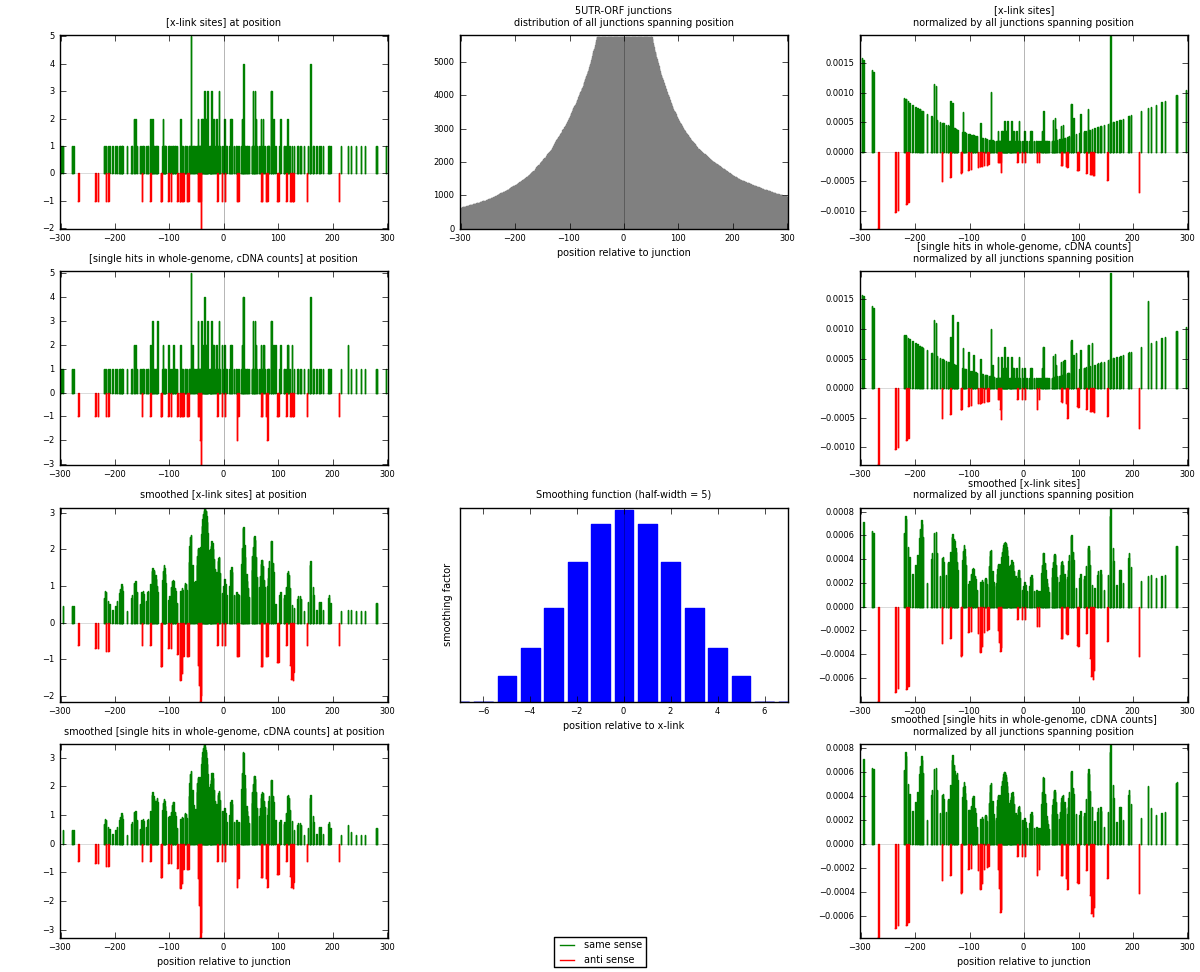 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 241 | 0.18% | 0.56% | 85.46% | 181 | 1.205 |
| anti | 41 | 0.03% | 0.10% | 14.54% | 21 | 0.205 |
| both | 282 | 0.21% | 0.65% | 100.00% | 200 | 1.41 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 259 | 0.18% | 0.53% | 85.20% | 181 | 1.295 |
| anti | 45 | 0.03% | 0.09% | 14.80% | 21 | 0.225 |
| both | 304 | 0.21% | 0.62% | 100.00% | 200 | 1.52 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
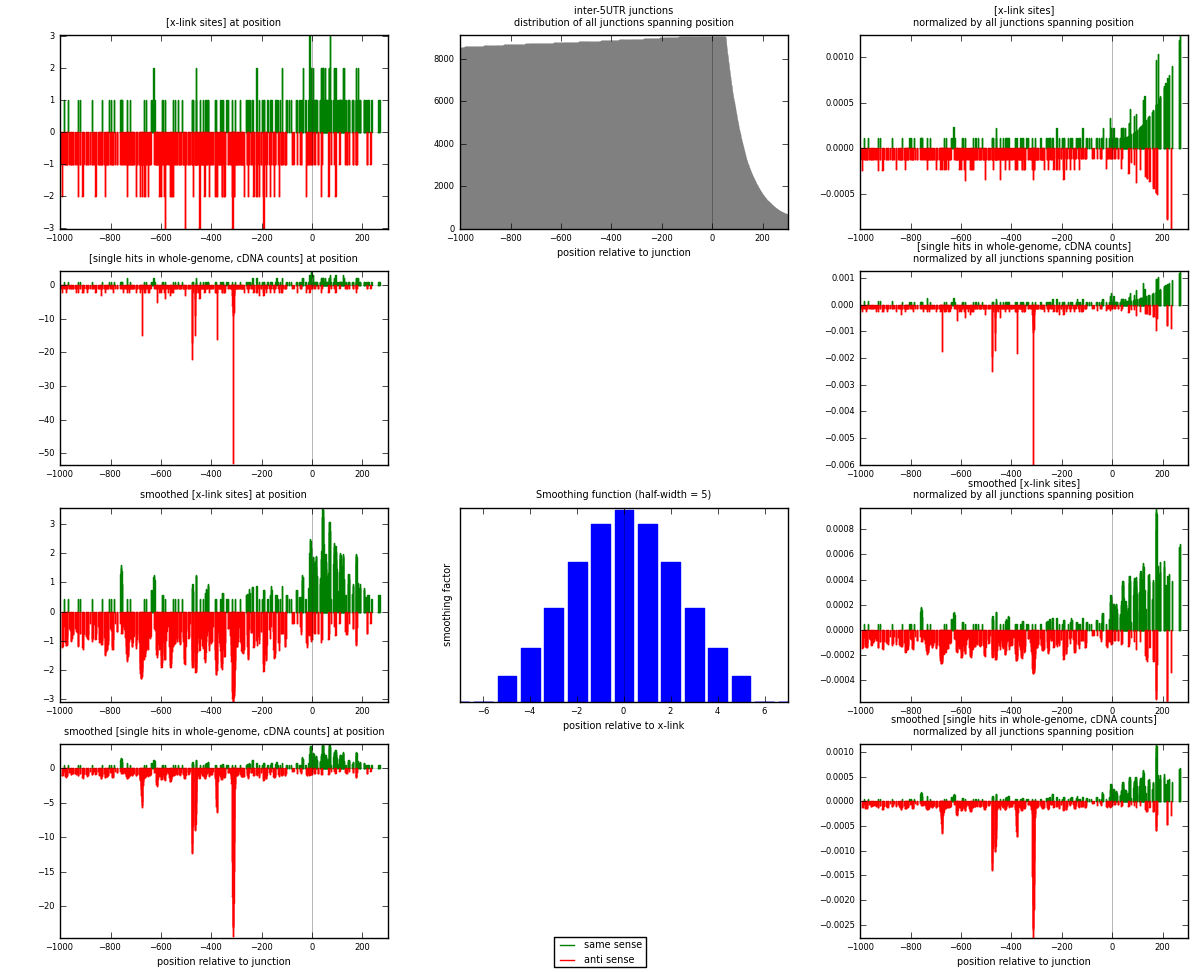 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 196 | 0.15% | 0.45% | 32.83% | 154 | 0.453704 |
| anti | 401 | 0.30% | 0.93% | 67.17% | 289 | 0.928241 |
| both | 597 | 0.45% | 1.39% | 100.00% | 432 | 1.38194 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 207 | 0.14% | 0.42% | 25.71% | 154 | 0.479167 |
| anti | 598 | 0.41% | 1.21% | 74.29% | 289 | 1.38426 |
| both | 805 | 0.55% | 1.63% | 100.00% | 432 | 1.86343 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
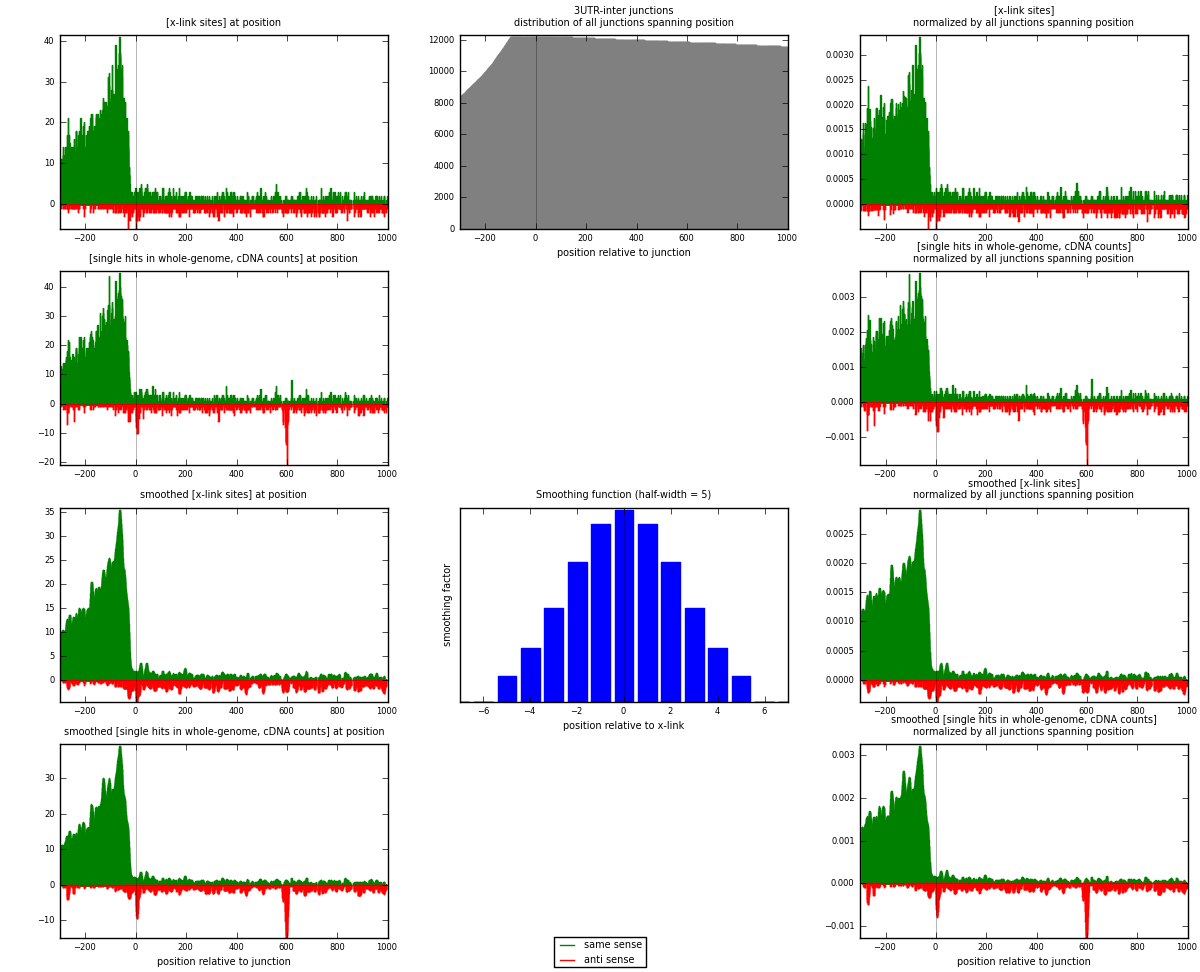 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5627 | 4.22% | 13.06% | 85.27% | 2419 | 2.05365 |
| anti | 972 | 0.73% | 2.26% | 14.73% | 405 | 0.354745 |
| both | 6599 | 4.95% | 15.31% | 100.00% | 2740 | 2.40839 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 6279 | 4.26% | 12.75% | 83.83% | 2419 | 2.29161 |
| anti | 1211 | 0.82% | 2.46% | 16.17% | 405 | 0.441971 |
| both | 7490 | 5.08% | 15.21% | 100.00% | 2740 | 2.73358 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
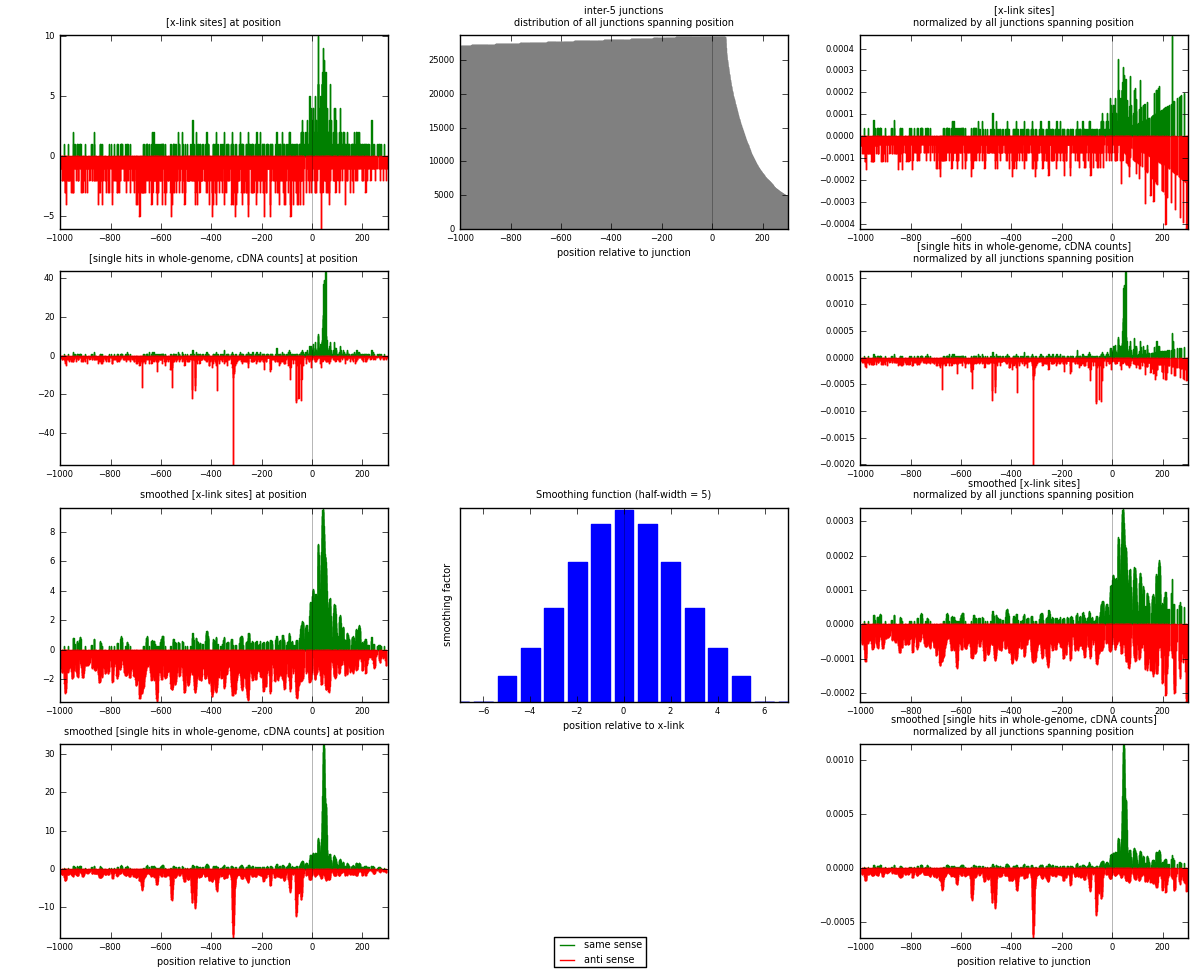 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 605 | 0.45% | 1.40% | 28.70% | 416 | 0.477883 |
| anti | 1503 | 1.13% | 3.49% | 71.30% | 870 | 1.1872 |
| both | 2108 | 1.58% | 4.89% | 100.00% | 1266 | 1.66509 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 864 | 0.59% | 1.75% | 30.56% | 416 | 0.682464 |
| anti | 1963 | 1.33% | 3.99% | 69.44% | 870 | 1.55055 |
| both | 2827 | 1.92% | 5.74% | 100.00% | 1266 | 2.23302 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
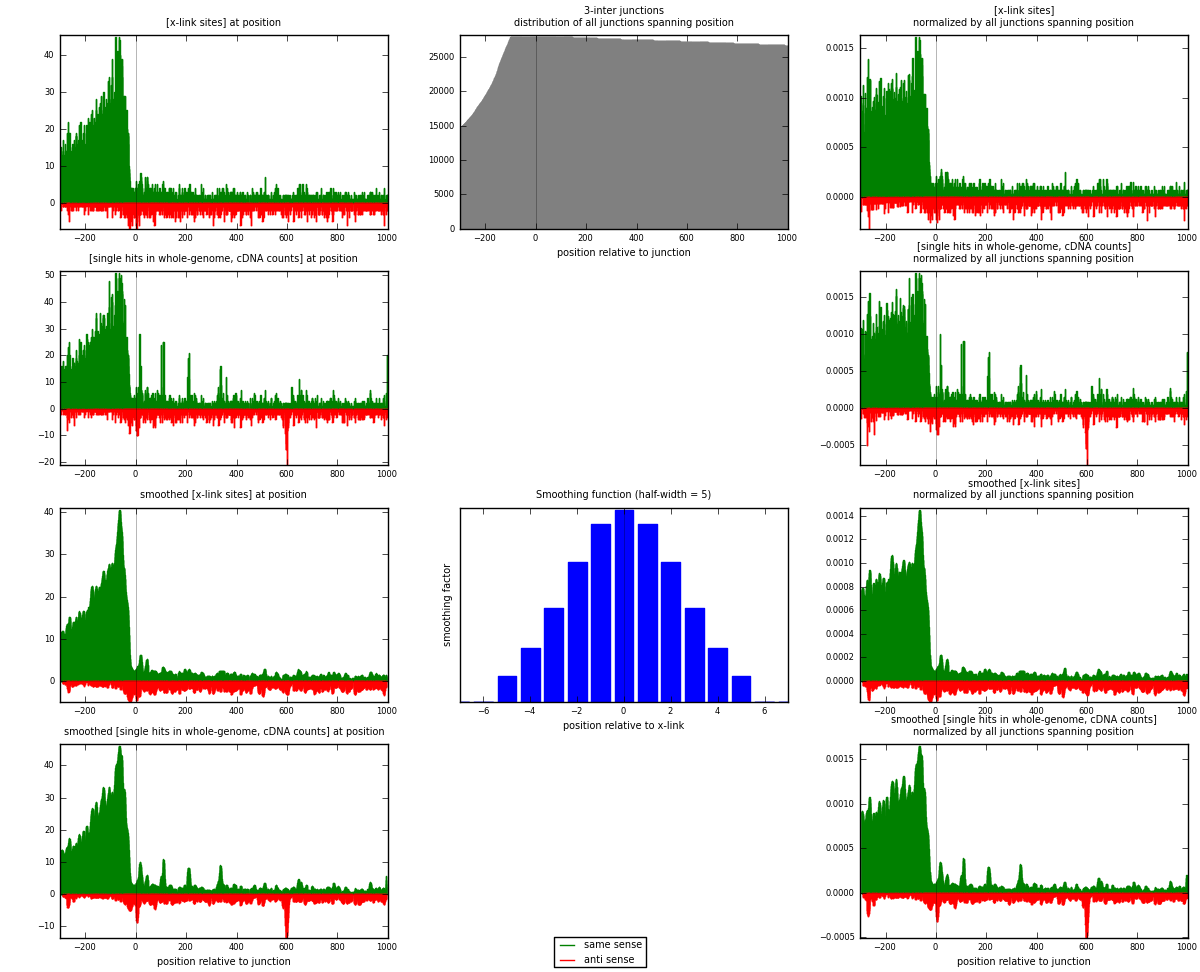 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6967 | 5.23% | 16.17% | 79.64% | 2946 | 1.90355 |
| anti | 1781 | 1.34% | 4.13% | 20.36% | 817 | 0.486612 |
| both | 8748 | 6.56% | 20.30% | 100.00% | 3660 | 2.39016 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 8363 | 5.67% | 16.98% | 79.88% | 2946 | 2.28497 |
| anti | 2107 | 1.43% | 4.28% | 20.12% | 817 | 0.575683 |
| both | 10470 | 7.10% | 21.26% | 100.00% | 3660 | 2.86066 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
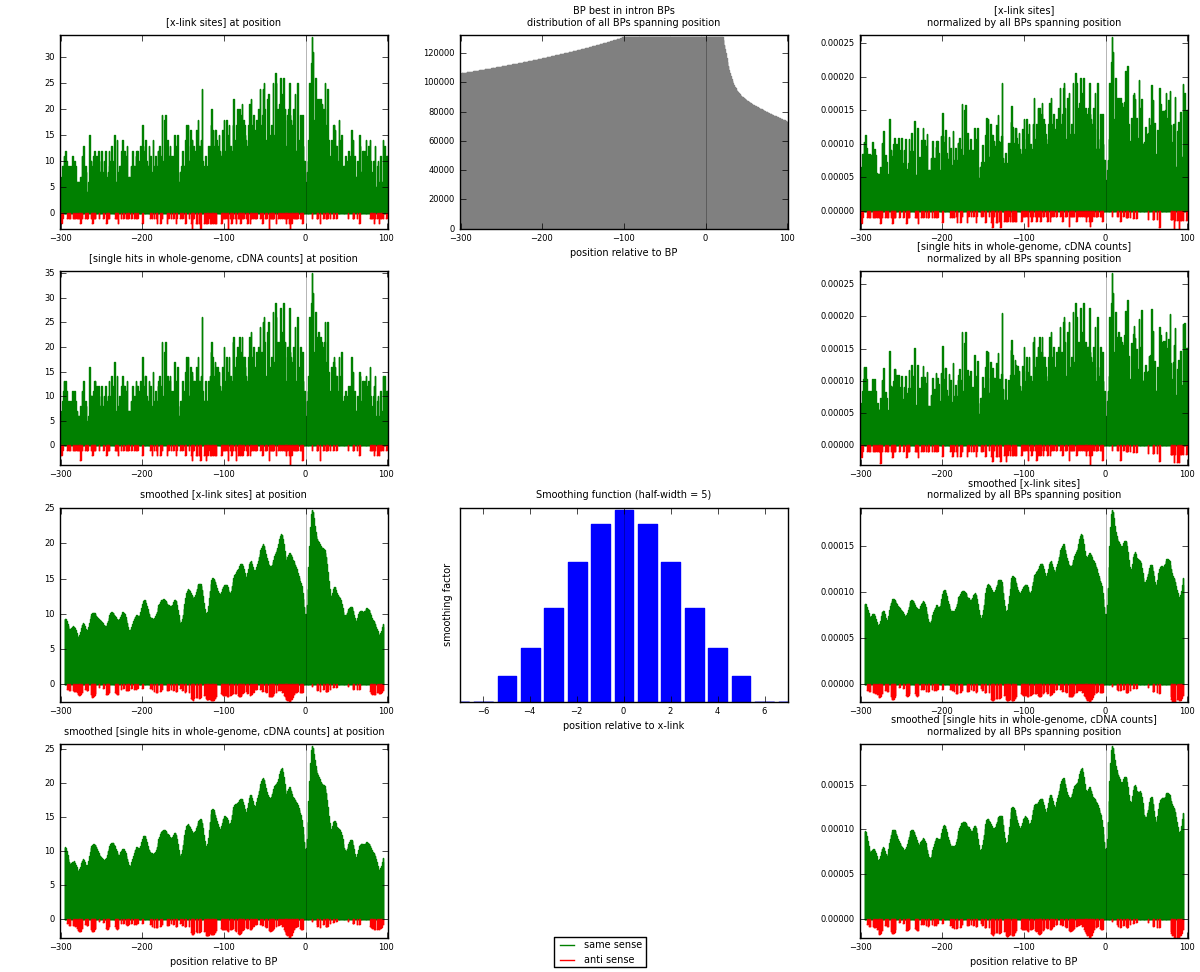 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5014 | 3.76% | 11.64% | 95.94% | 4381 | 1.10125 |
| anti | 212 | 0.16% | 0.49% | 4.06% | 179 | 0.0465627 |
| both | 5226 | 3.92% | 12.13% | 100.00% | 4553 | 1.14781 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 5240 | 3.55% | 10.64% | 95.94% | 4381 | 1.15089 |
| anti | 222 | 0.15% | 0.45% | 4.06% | 179 | 0.0487591 |
| both | 5462 | 3.70% | 11.09% | 100.00% | 4553 | 1.19965 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.