RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
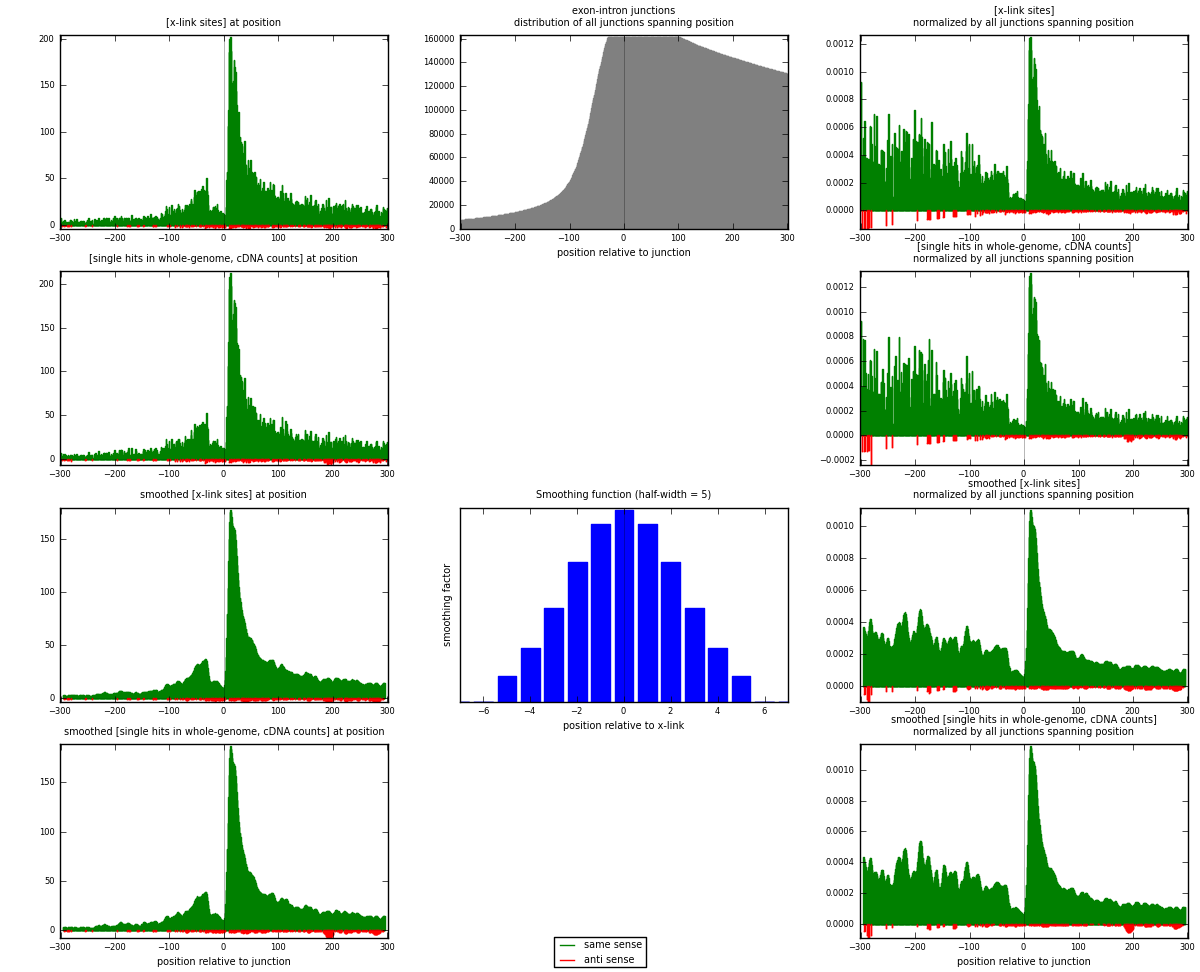 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13615 | 9.55% | 28.95% | 97.82% | 10664 | 1.25357 |
| anti | 304 | 0.21% | 0.65% | 2.18% | 213 | 0.0279901 |
| both | 13919 | 9.76% | 29.59% | 100.00% | 10861 | 1.28156 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 14218 | 9.18% | 27.14% | 97.49% | 10664 | 1.30909 |
| anti | 366 | 0.24% | 0.70% | 2.51% | 213 | 0.0336986 |
| both | 14584 | 9.42% | 27.83% | 100.00% | 10861 | 1.34279 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
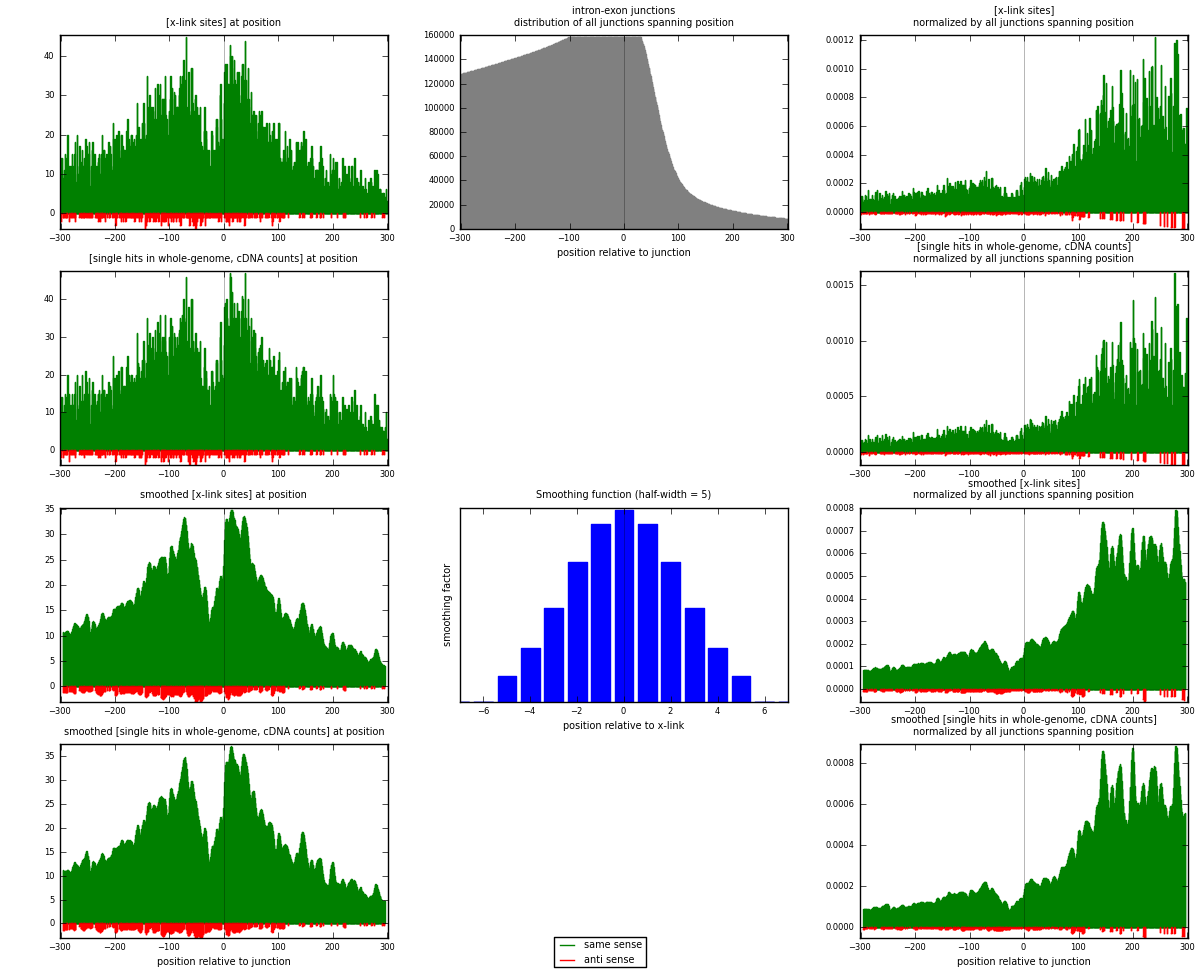 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9940 | 6.97% | 21.13% | 96.82% | 7693 | 1.25063 |
| anti | 326 | 0.23% | 0.69% | 3.18% | 277 | 0.0410166 |
| both | 10266 | 7.20% | 21.83% | 100.00% | 7948 | 1.29165 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 10534 | 6.80% | 20.10% | 96.89% | 7693 | 1.32536 |
| anti | 338 | 0.22% | 0.65% | 3.11% | 277 | 0.0425264 |
| both | 10872 | 7.02% | 20.75% | 100.00% | 7948 | 1.36789 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
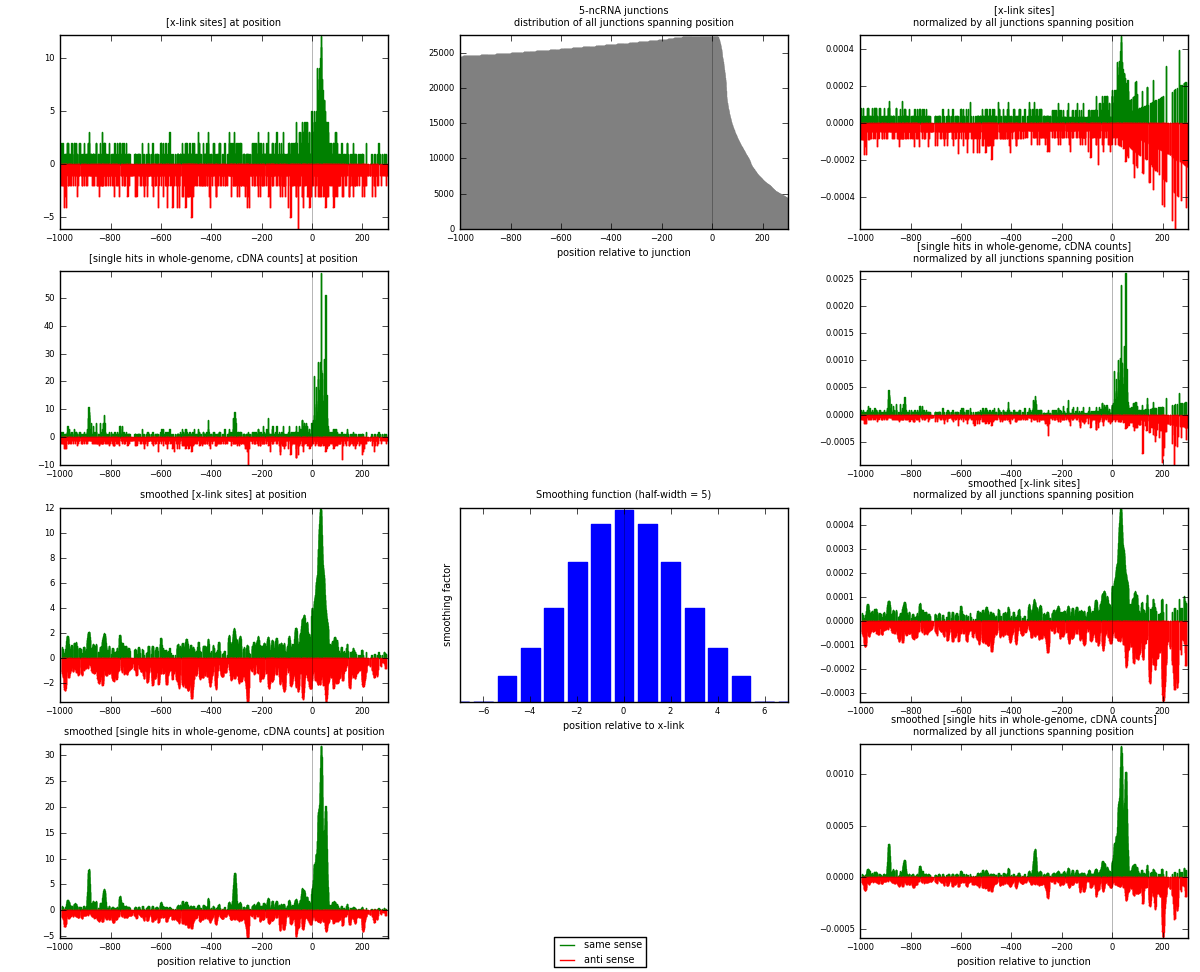 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1004 | 0.70% | 2.13% | 45.16% | 482 | 0.886143 |
| anti | 1219 | 0.86% | 2.59% | 54.84% | 659 | 1.0759 |
| both | 2223 | 1.56% | 4.73% | 100.00% | 1133 | 1.96205 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1663 | 1.07% | 3.17% | 55.29% | 482 | 1.46778 |
| anti | 1345 | 0.87% | 2.57% | 44.71% | 659 | 1.18711 |
| both | 3008 | 1.94% | 5.74% | 100.00% | 1133 | 2.6549 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
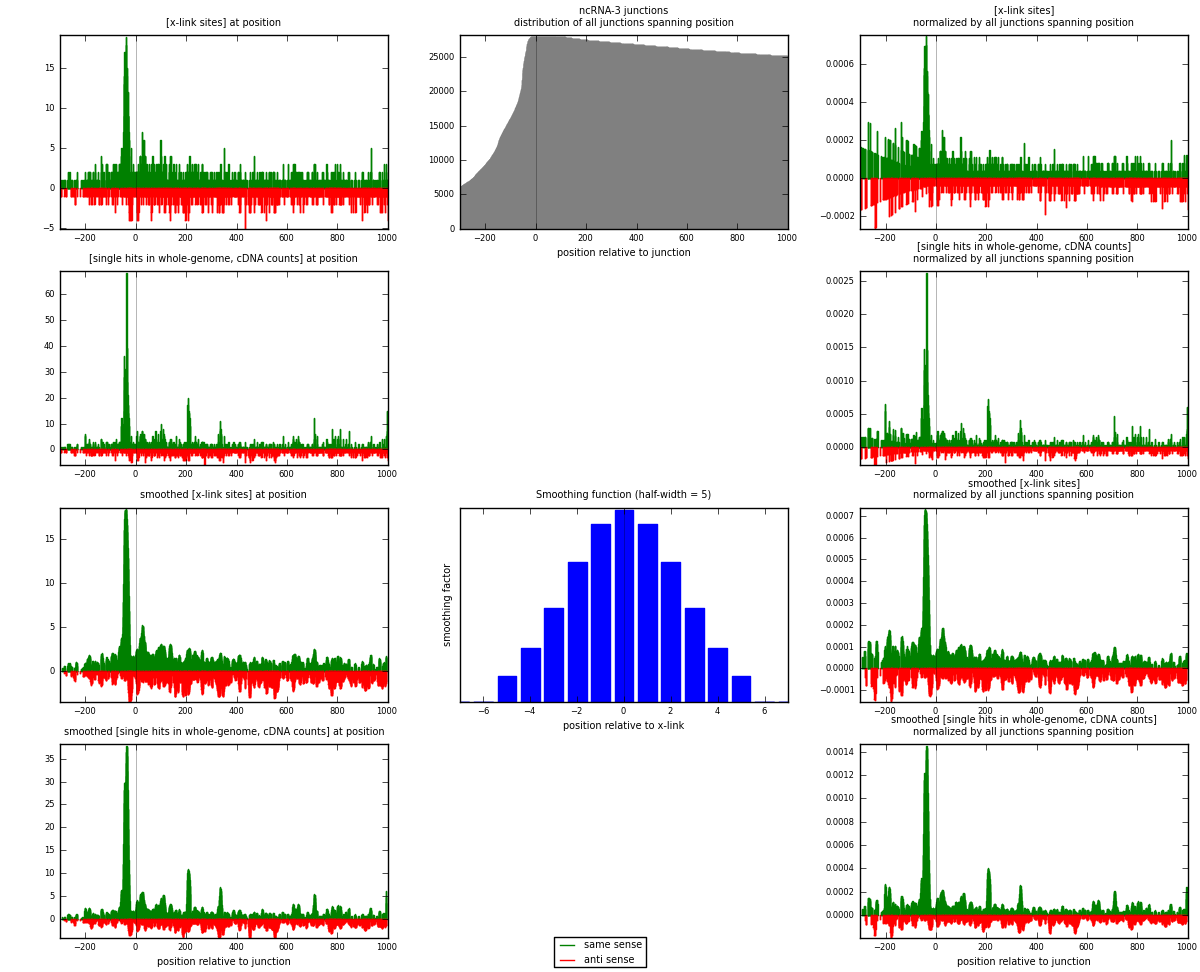 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1474 | 1.03% | 3.13% | 60.14% | 708 | 1.18204 |
| anti | 977 | 0.69% | 2.08% | 39.86% | 552 | 0.78348 |
| both | 2451 | 1.72% | 5.21% | 100.00% | 1247 | 1.96552 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2138 | 1.38% | 4.08% | 66.73% | 708 | 1.71451 |
| anti | 1066 | 0.69% | 2.03% | 33.27% | 552 | 0.854852 |
| both | 3204 | 2.07% | 6.11% | 100.00% | 1247 | 2.56937 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
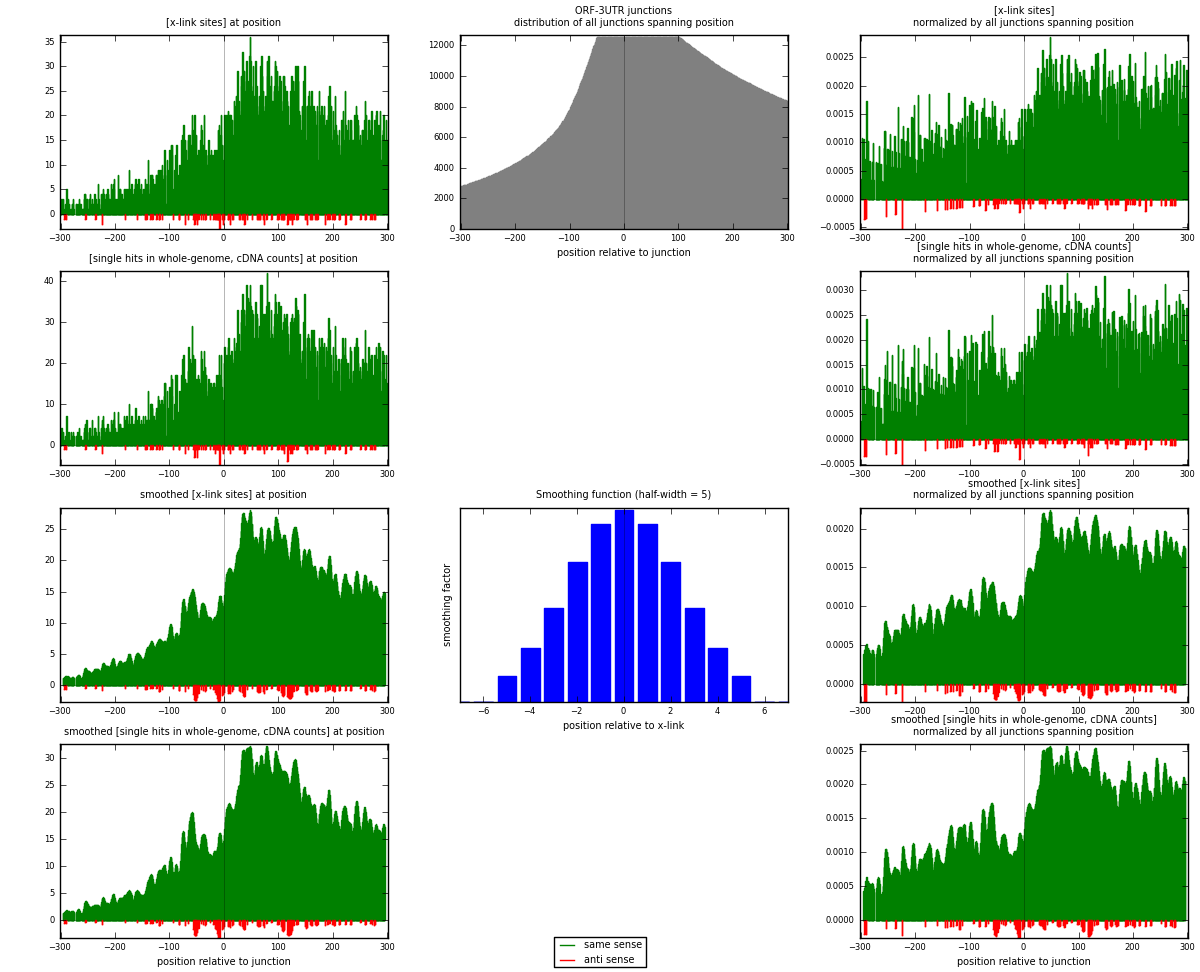
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7821 | 5.49% | 16.63% | 98.45% | 2607 | 2.92702 |
| anti | 123 | 0.09% | 0.26% | 1.55% | 75 | 0.0460329 |
| both | 7944 | 5.57% | 16.89% | 100.00% | 2672 | 2.97305 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9111 | 5.89% | 17.39% | 98.57% | 2607 | 3.40981 |
| anti | 132 | 0.09% | 0.25% | 1.43% | 75 | 0.0494012 |
| both | 9243 | 5.97% | 17.64% | 100.00% | 2672 | 3.45921 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
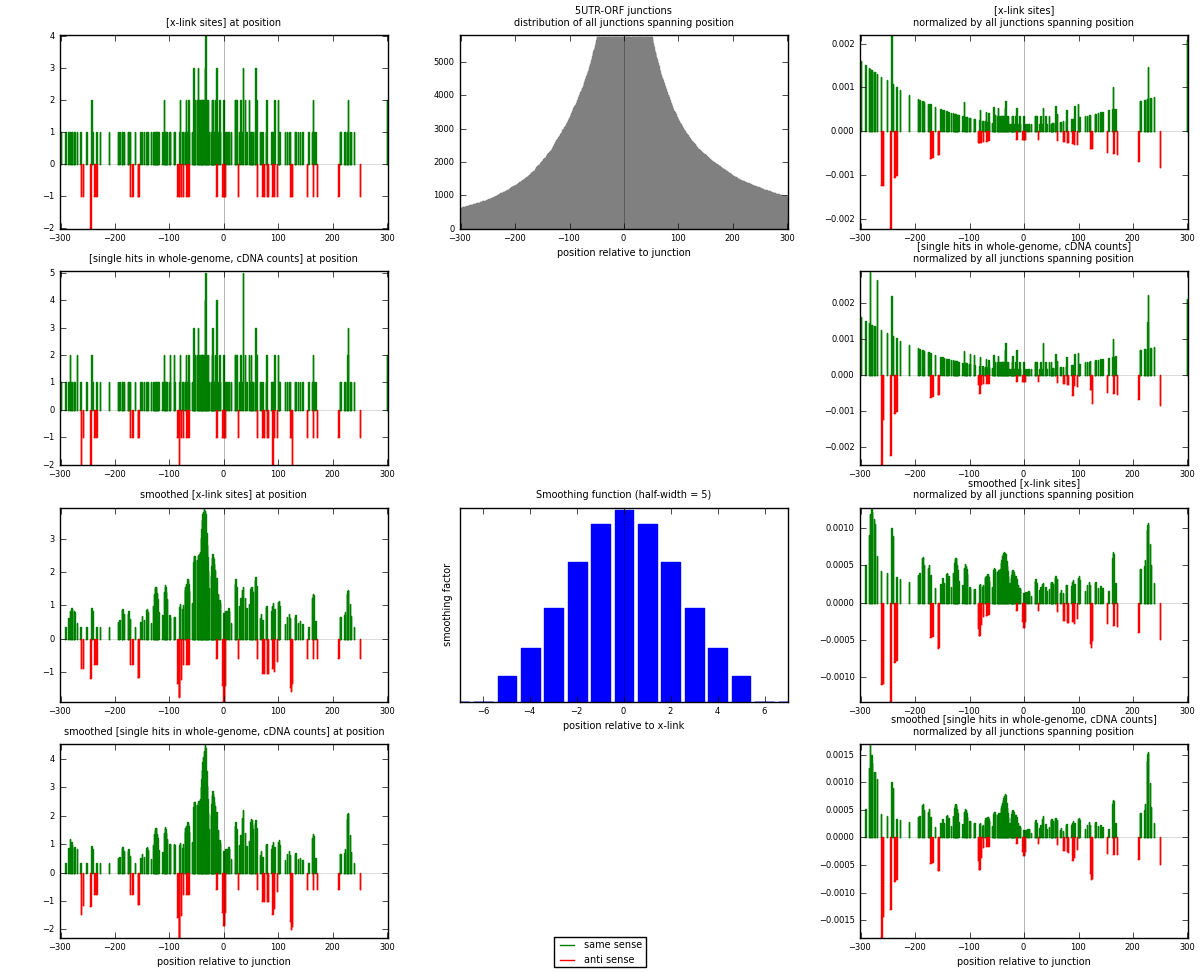
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 207 | 0.15% | 0.44% | 84.15% | 145 | 1.29375 |
| anti | 39 | 0.03% | 0.08% | 15.85% | 16 | 0.24375 |
| both | 246 | 0.17% | 0.52% | 100.00% | 160 | 1.5375 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 220 | 0.14% | 0.42% | 83.65% | 145 | 1.375 |
| anti | 43 | 0.03% | 0.08% | 16.35% | 16 | 0.26875 |
| both | 263 | 0.17% | 0.50% | 100.00% | 160 | 1.64375 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
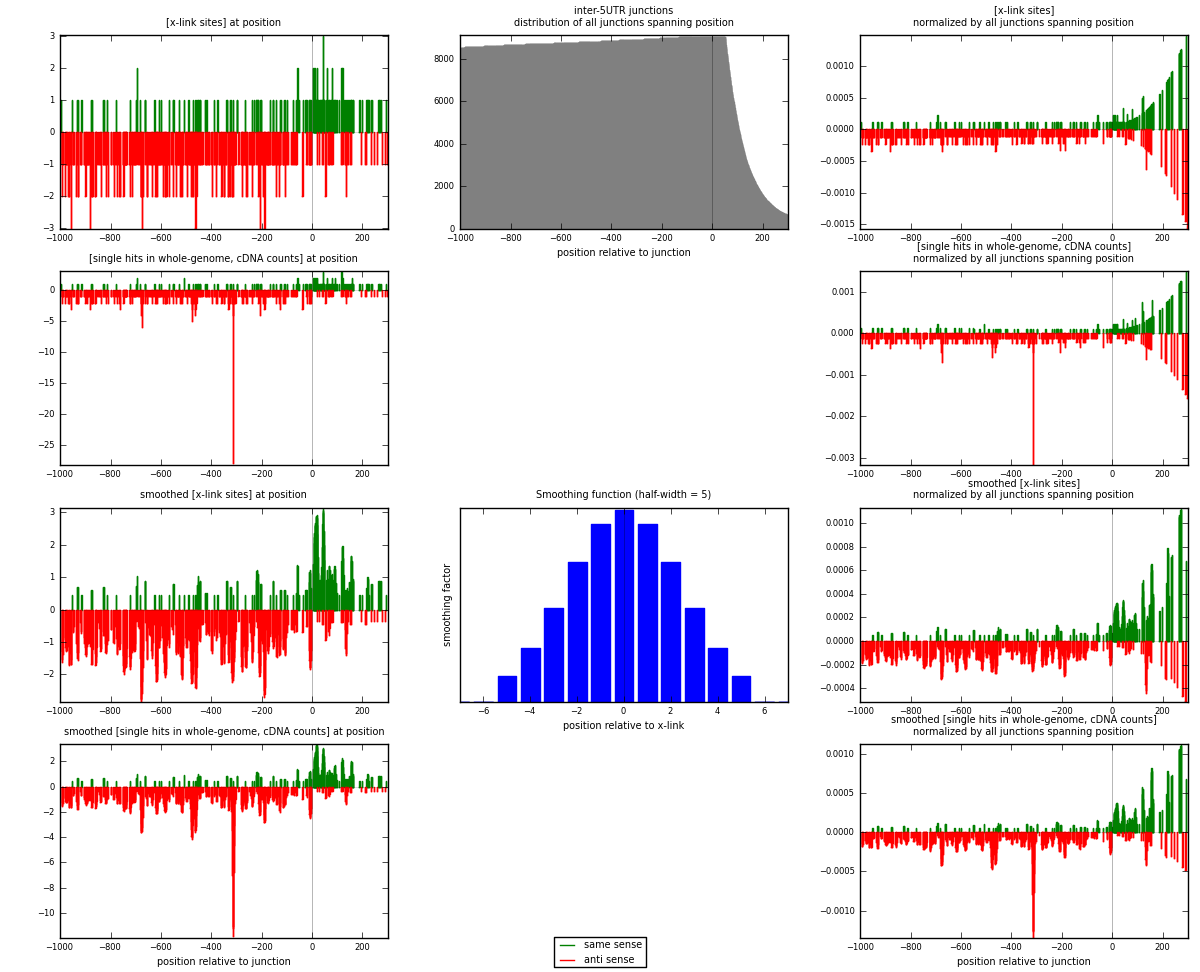
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 161 | 0.11% | 0.34% | 25.84% | 129 | 0.378824 |
| anti | 462 | 0.32% | 0.98% | 74.16% | 306 | 1.08706 |
| both | 623 | 0.44% | 1.32% | 100.00% | 425 | 1.46588 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 166 | 0.11% | 0.32% | 23.88% | 129 | 0.390588 |
| anti | 529 | 0.34% | 1.01% | 76.12% | 306 | 1.24471 |
| both | 695 | 0.45% | 1.33% | 100.00% | 425 | 1.63529 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
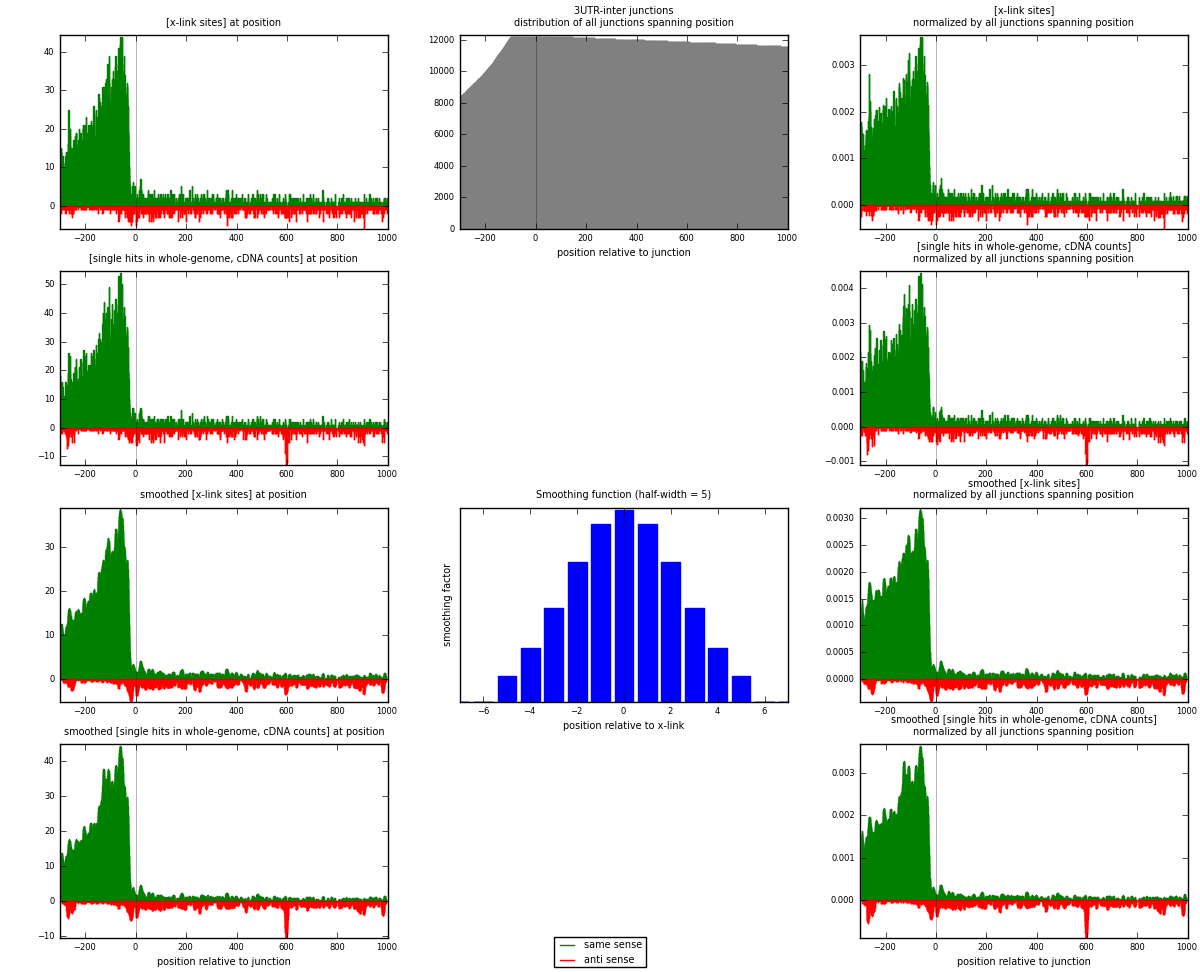
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 6529 | 4.58% | 13.88% | 85.67% | 2527 | 2.26938 |
| anti | 1092 | 0.77% | 2.32% | 14.33% | 423 | 0.379562 |
| both | 7621 | 5.35% | 16.20% | 100.00% | 2877 | 2.64894 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 7354 | 4.75% | 14.04% | 85.15% | 2527 | 2.55613 |
| anti | 1283 | 0.83% | 2.45% | 14.85% | 423 | 0.445951 |
| both | 8637 | 5.58% | 16.48% | 100.00% | 2877 | 3.00209 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
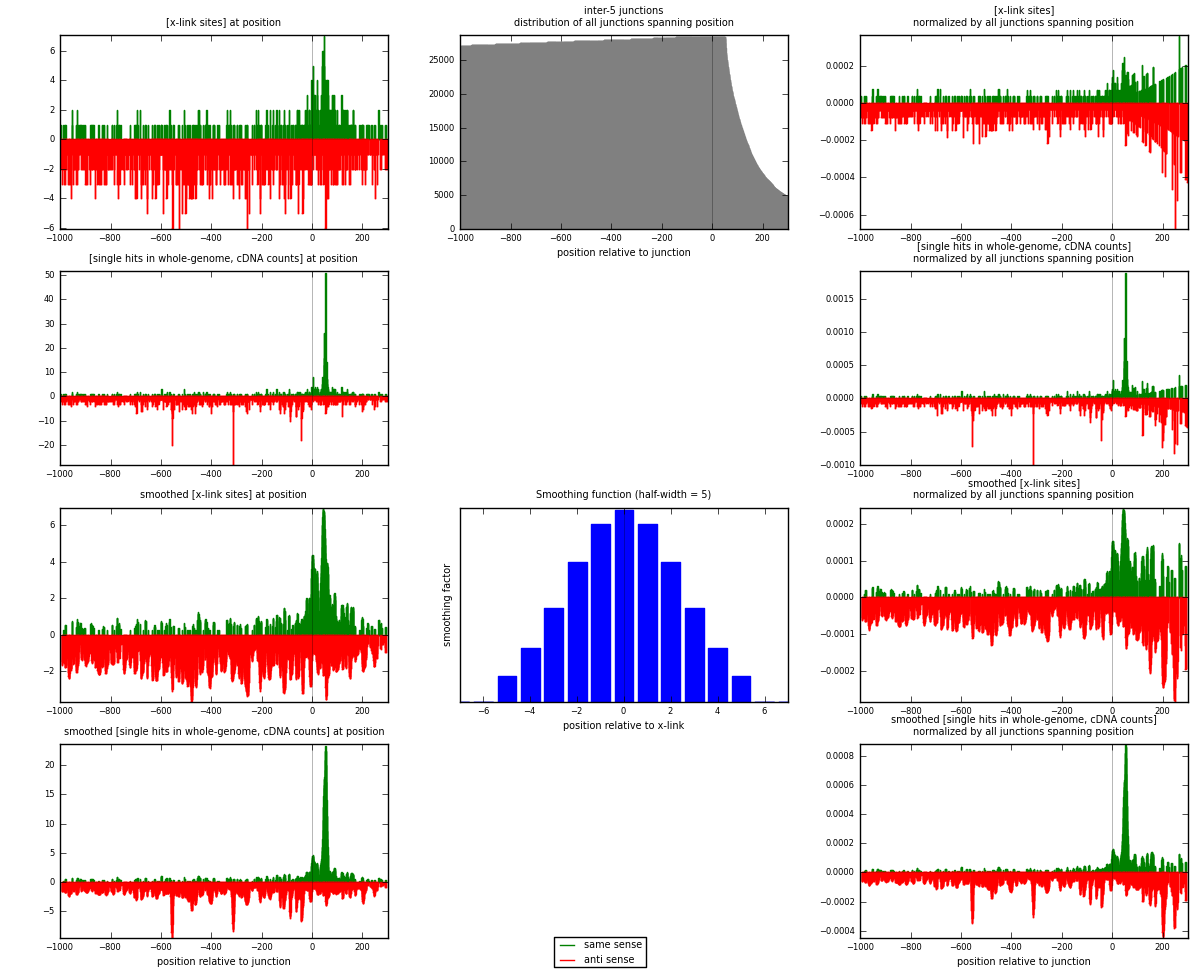 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 472 | 0.33% | 1.00% | 23.26% | 312 | 0.408658 |
| anti | 1557 | 1.09% | 3.31% | 76.74% | 860 | 1.34805 |
| both | 2029 | 1.42% | 4.31% | 100.00% | 1155 | 1.75671 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 654 | 0.42% | 1.25% | 26.24% | 312 | 0.566234 |
| anti | 1838 | 1.19% | 3.51% | 73.76% | 860 | 1.59134 |
| both | 2492 | 1.61% | 4.76% | 100.00% | 1155 | 2.15758 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
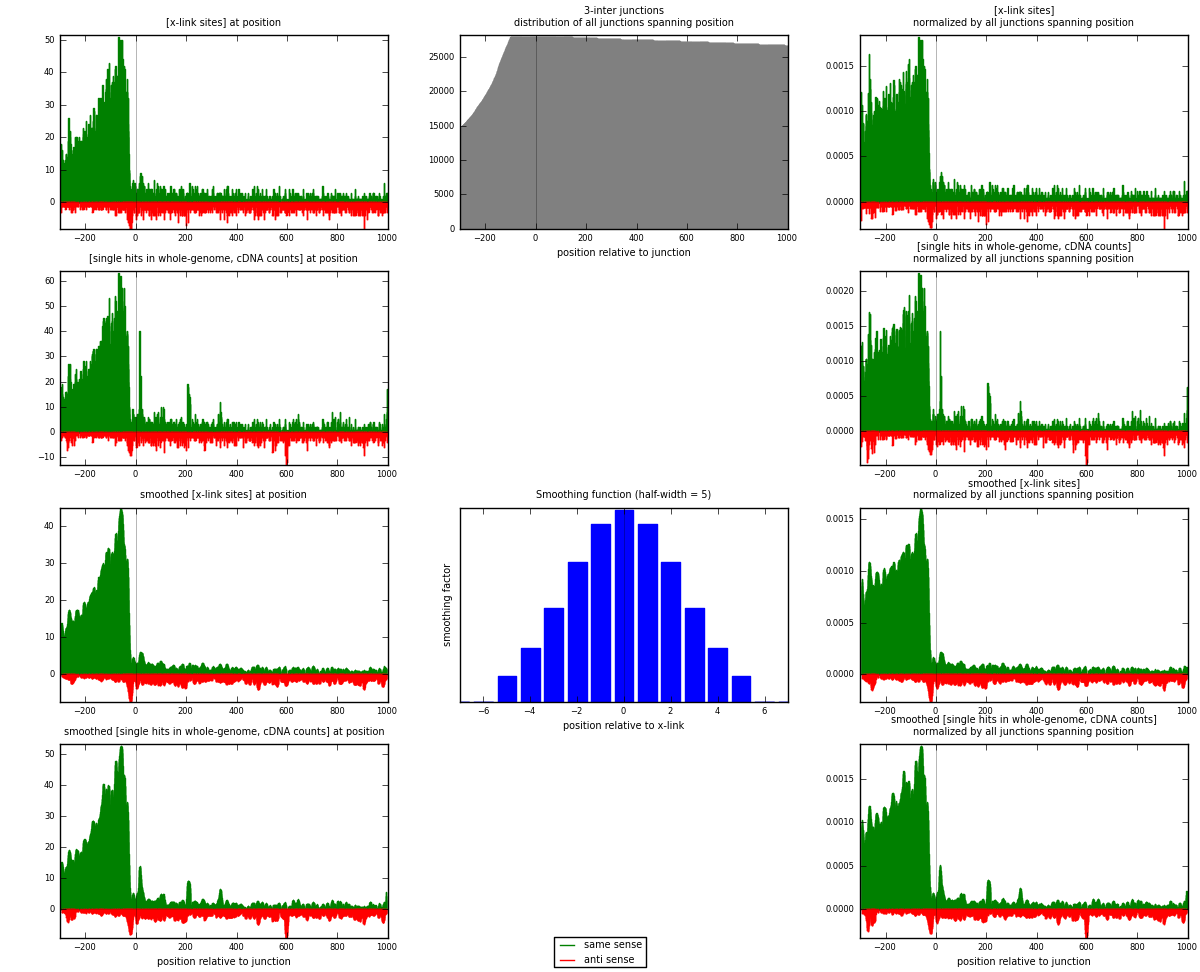 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 7959 | 5.58% | 16.92% | 80.74% | 3059 | 2.10779 |
| anti | 1899 | 1.33% | 4.04% | 19.26% | 807 | 0.502913 |
| both | 9858 | 6.91% | 20.96% | 100.00% | 3776 | 2.6107 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9430 | 6.09% | 18.00% | 81.08% | 3059 | 2.49735 |
| anti | 2200 | 1.42% | 4.20% | 18.92% | 807 | 0.582627 |
| both | 11630 | 7.51% | 22.20% | 100.00% | 3776 | 3.07998 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
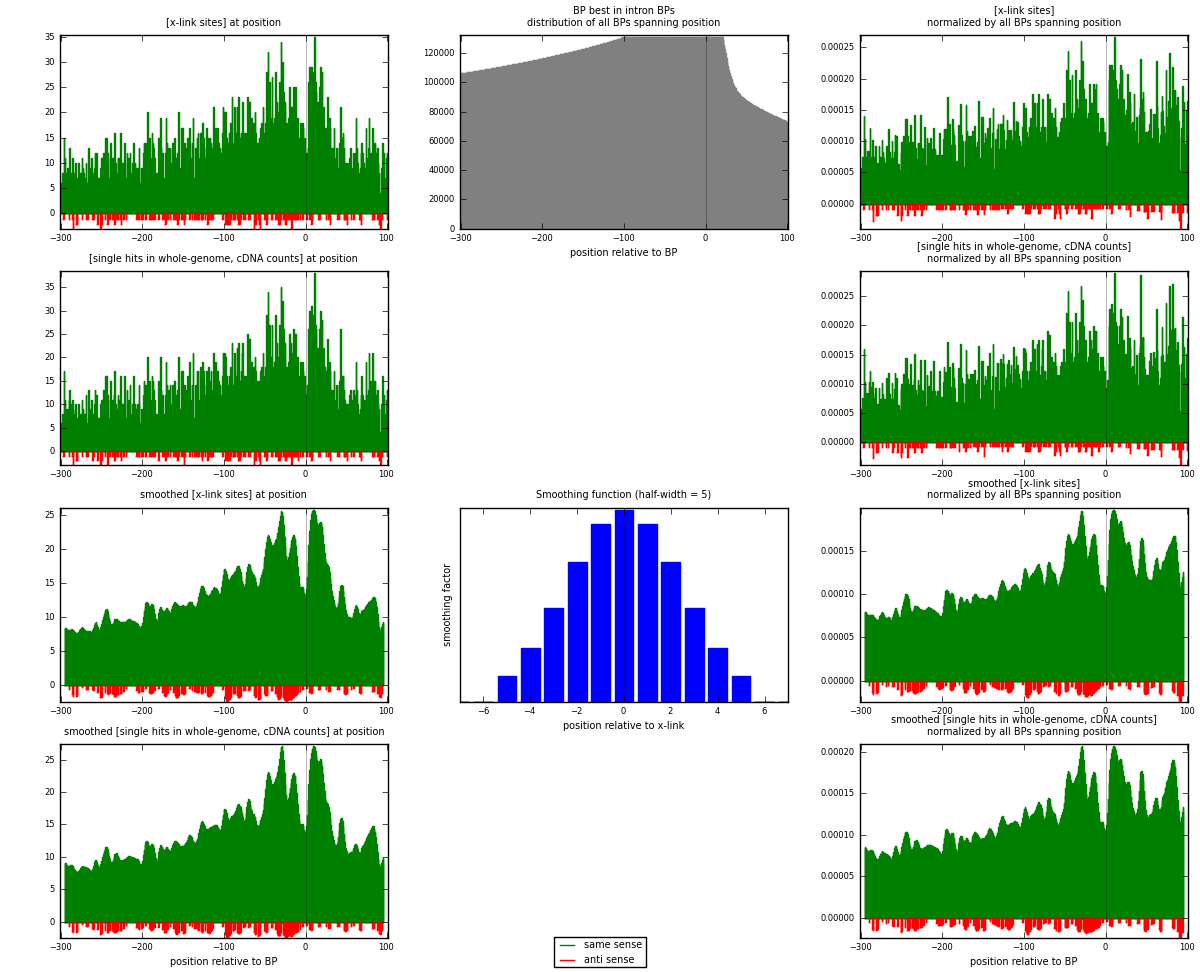 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 5357 | 3.76% | 11.39% | 96.52% | 4570 | 1.13352 |
| anti | 193 | 0.14% | 0.41% | 3.48% | 160 | 0.0408379 |
| both | 5550 | 3.89% | 11.80% | 100.00% | 4726 | 1.17435 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 5577 | 3.60% | 10.64% | 96.55% | 4570 | 1.18007 |
| anti | 199 | 0.13% | 0.38% | 3.45% | 160 | 0.0421075 |
| both | 5776 | 3.73% | 11.02% | 100.00% | 4726 | 1.22218 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.