RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
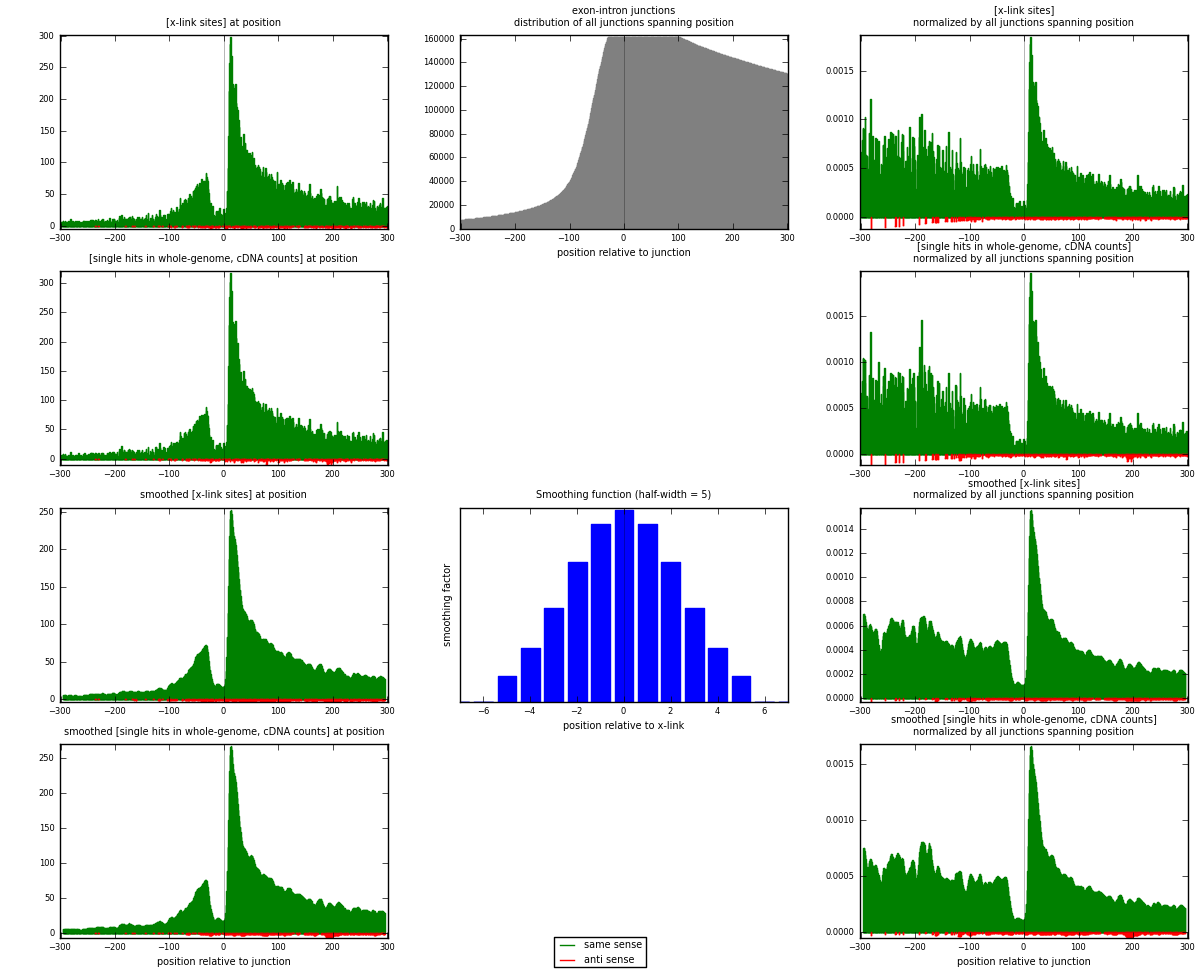 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 24520 | 9.31% | 30.90% | 97.64% | 17296 | 1.38829 |
| anti | 593 | 0.23% | 0.75% | 2.36% | 422 | 0.0335749 |
| both | 25113 | 9.54% | 31.65% | 100.00% | 17662 | 1.42187 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 25686 | 8.93% | 29.06% | 97.35% | 17296 | 1.45431 |
| anti | 699 | 0.24% | 0.79% | 2.65% | 422 | 0.0395765 |
| both | 26385 | 9.18% | 29.85% | 100.00% | 17662 | 1.49389 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
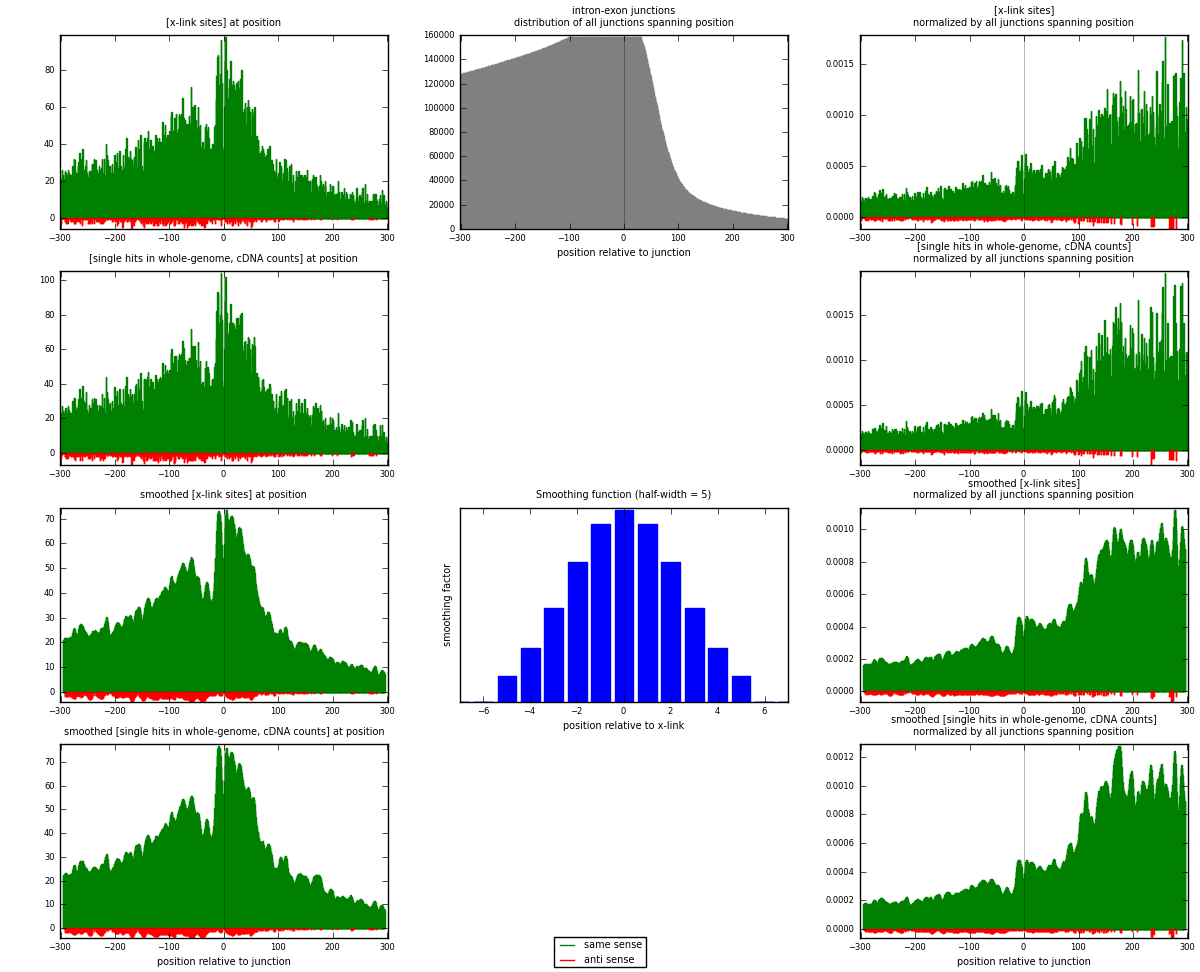 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 18365 | 6.97% | 23.15% | 96.80% | 13354 | 1.33051 |
| anti | 607 | 0.23% | 0.77% | 3.20% | 487 | 0.0439759 |
| both | 18972 | 7.20% | 23.91% | 100.00% | 13803 | 1.37448 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 19433 | 6.76% | 21.99% | 96.82% | 13354 | 1.40788 |
| anti | 639 | 0.22% | 0.72% | 3.18% | 487 | 0.0462943 |
| both | 20072 | 6.98% | 22.71% | 100.00% | 13803 | 1.45418 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
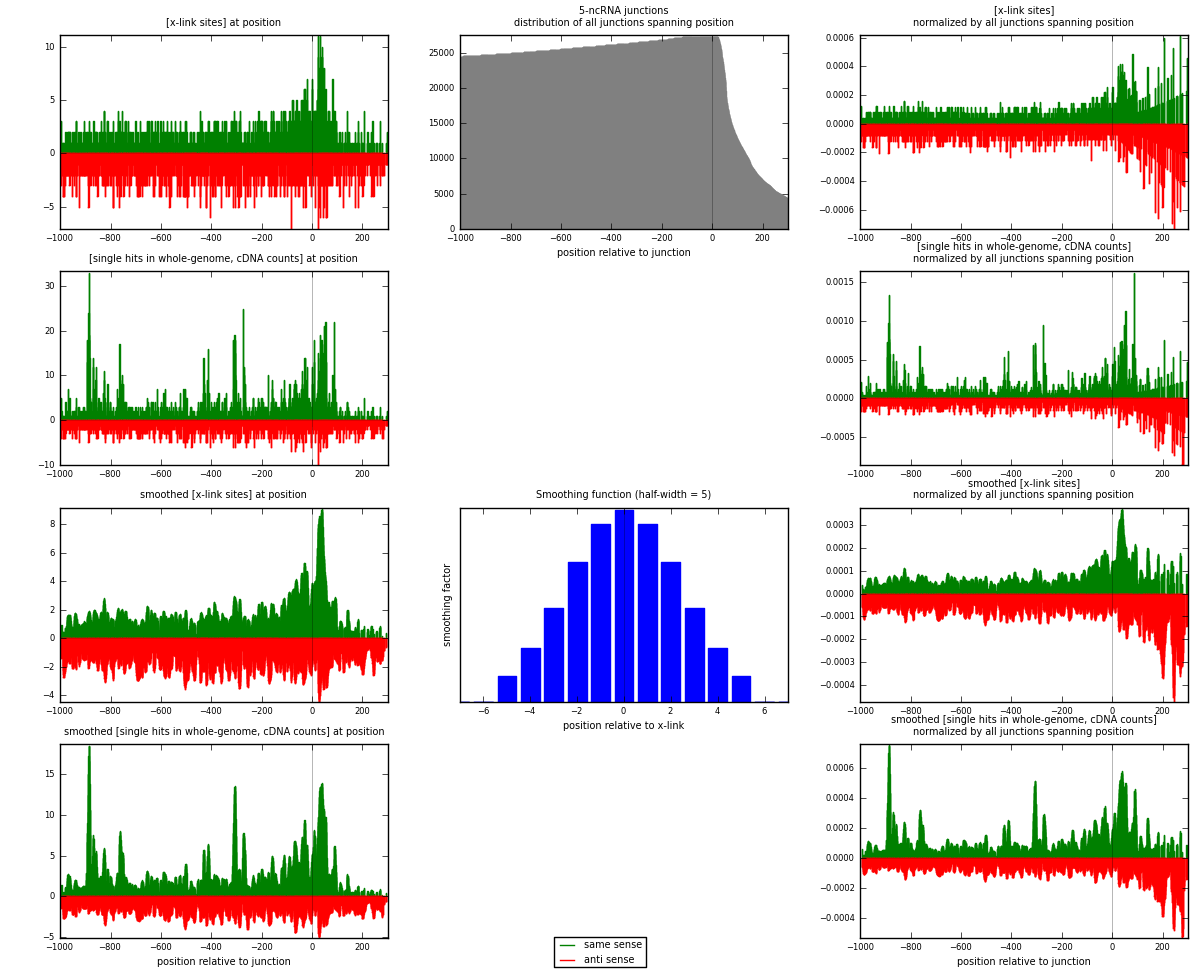 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1869 | 0.71% | 2.36% | 47.98% | 792 | 1.03776 |
| anti | 2026 | 0.77% | 2.55% | 52.02% | 1036 | 1.12493 |
| both | 3895 | 1.48% | 4.91% | 100.00% | 1801 | 2.16269 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3033 | 1.06% | 3.43% | 58.15% | 792 | 1.68406 |
| anti | 2183 | 0.76% | 2.47% | 41.85% | 1036 | 1.2121 |
| both | 5216 | 1.81% | 5.90% | 100.00% | 1801 | 2.89617 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
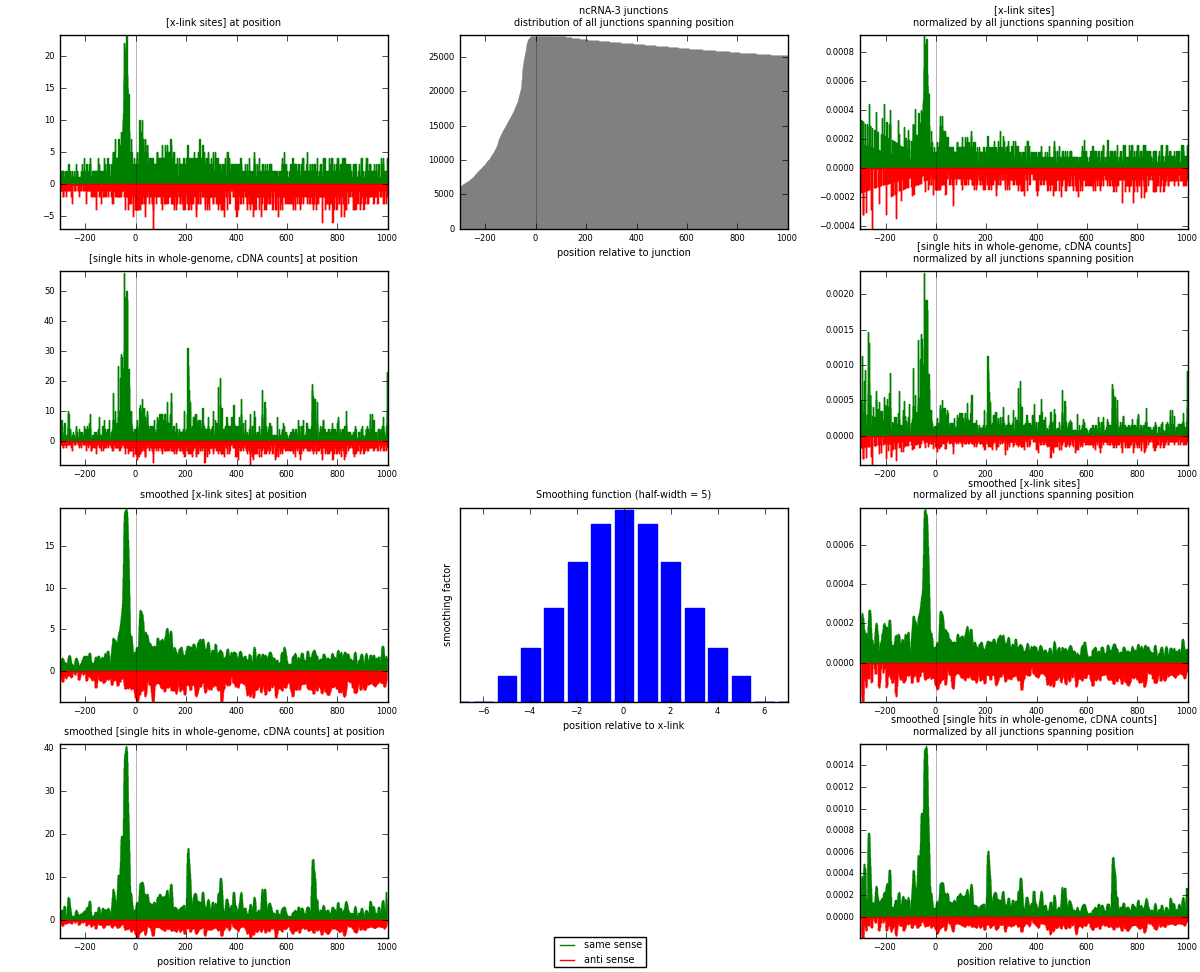 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2760 | 1.05% | 3.48% | 61.99% | 1164 | 1.35228 |
| anti | 1692 | 0.64% | 2.13% | 38.01% | 906 | 0.829005 |
| both | 4452 | 1.69% | 5.61% | 100.00% | 2041 | 2.18128 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4633 | 1.61% | 5.24% | 71.95% | 1164 | 2.26997 |
| anti | 1806 | 0.63% | 2.04% | 28.05% | 906 | 0.88486 |
| both | 6439 | 2.24% | 7.29% | 100.00% | 2041 | 3.15483 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
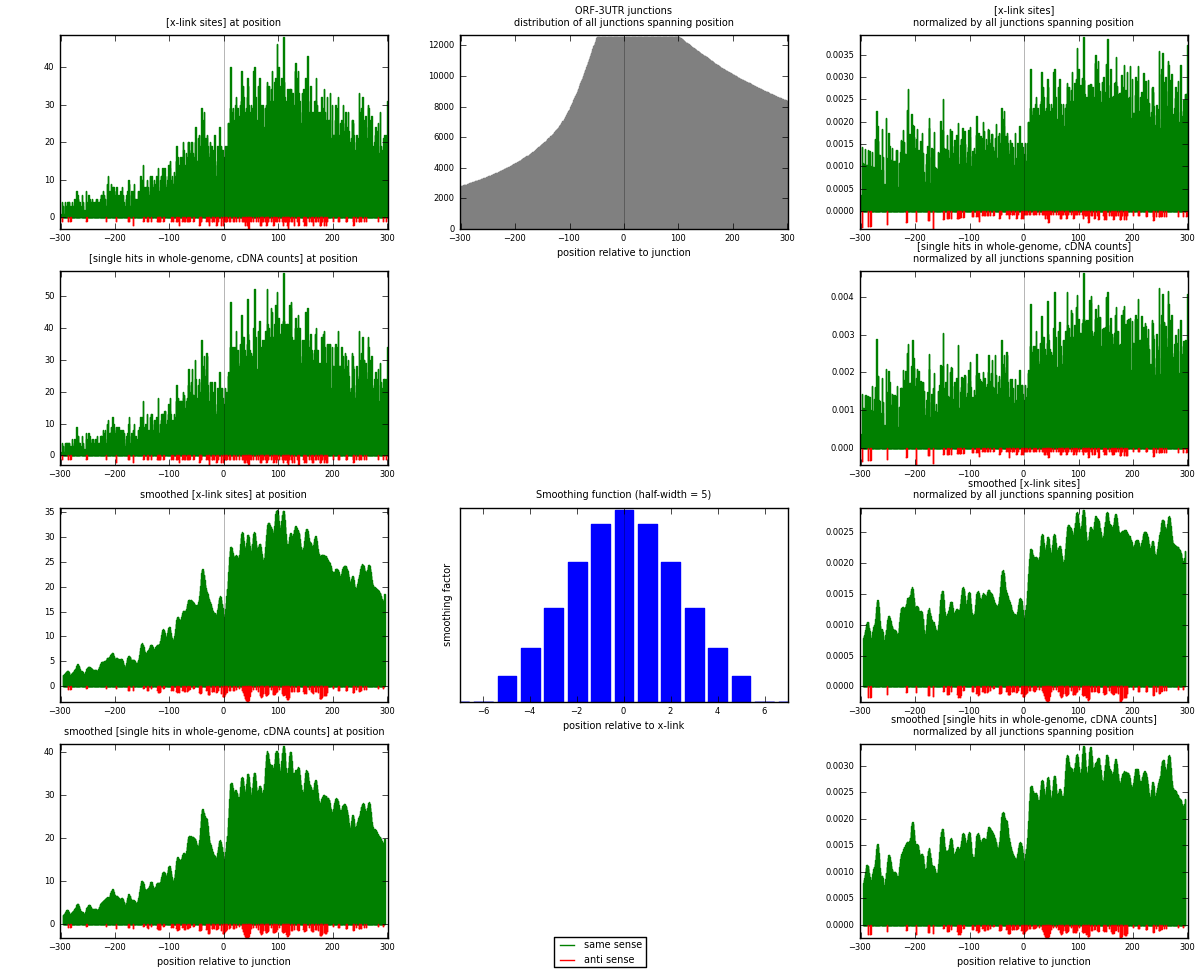 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 10550 | 4.01% | 13.30% | 98.46% | 3155 | 3.25216 |
| anti | 165 | 0.06% | 0.21% | 1.54% | 116 | 0.0508631 |
| both | 10715 | 4.07% | 13.50% | 100.00% | 3244 | 3.30302 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 12048 | 4.19% | 13.63% | 98.58% | 3155 | 3.71393 |
| anti | 173 | 0.06% | 0.20% | 1.42% | 116 | 0.0533292 |
| both | 12221 | 4.25% | 13.83% | 100.00% | 3244 | 3.76726 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
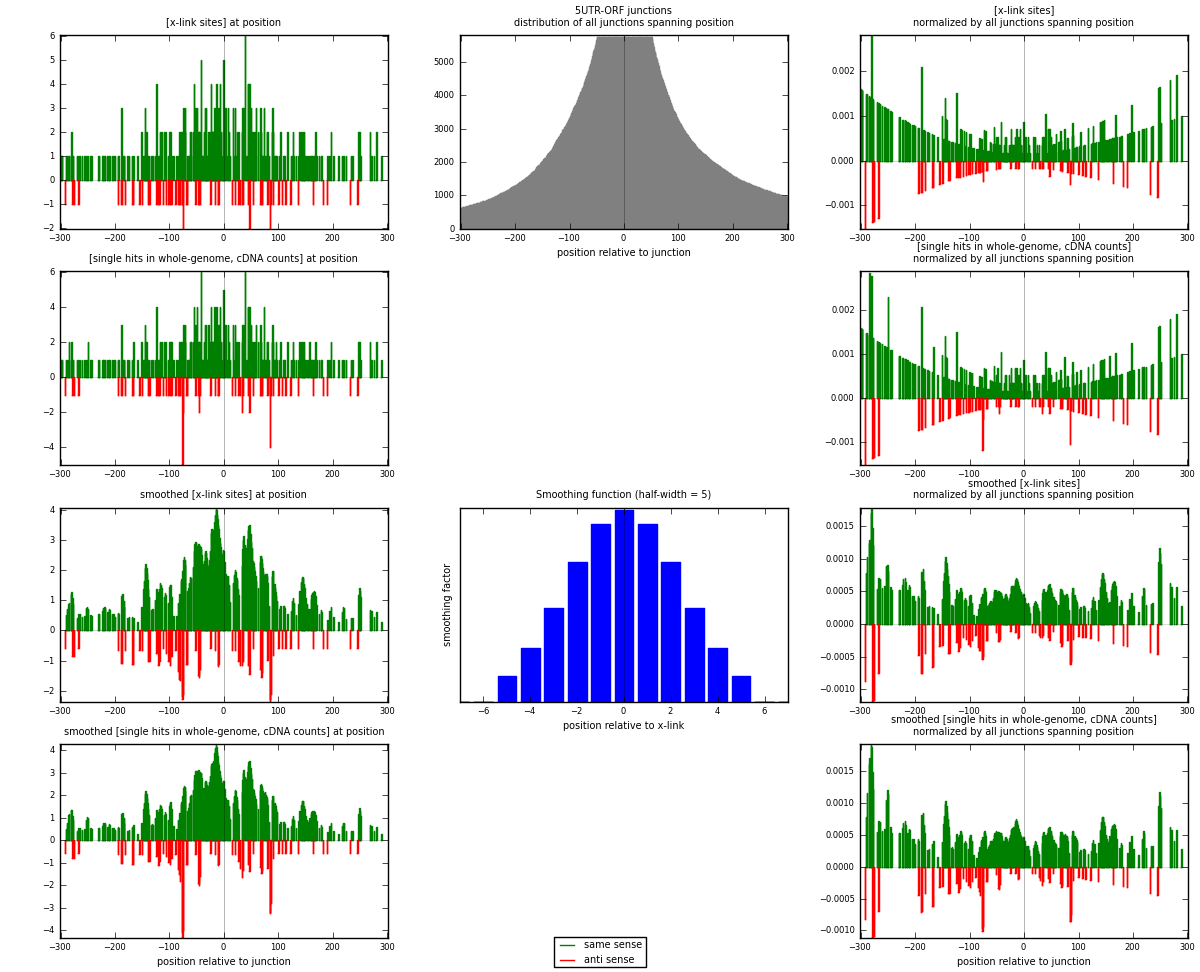 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 391 | 0.15% | 0.49% | 85.00% | 267 | 1.33447 |
| anti | 69 | 0.03% | 0.09% | 15.00% | 28 | 0.235495 |
| both | 460 | 0.17% | 0.58% | 100.00% | 293 | 1.56997 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 404 | 0.14% | 0.46% | 83.99% | 267 | 1.37884 |
| anti | 77 | 0.03% | 0.09% | 16.01% | 28 | 0.262799 |
| both | 481 | 0.17% | 0.54% | 100.00% | 293 | 1.64164 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
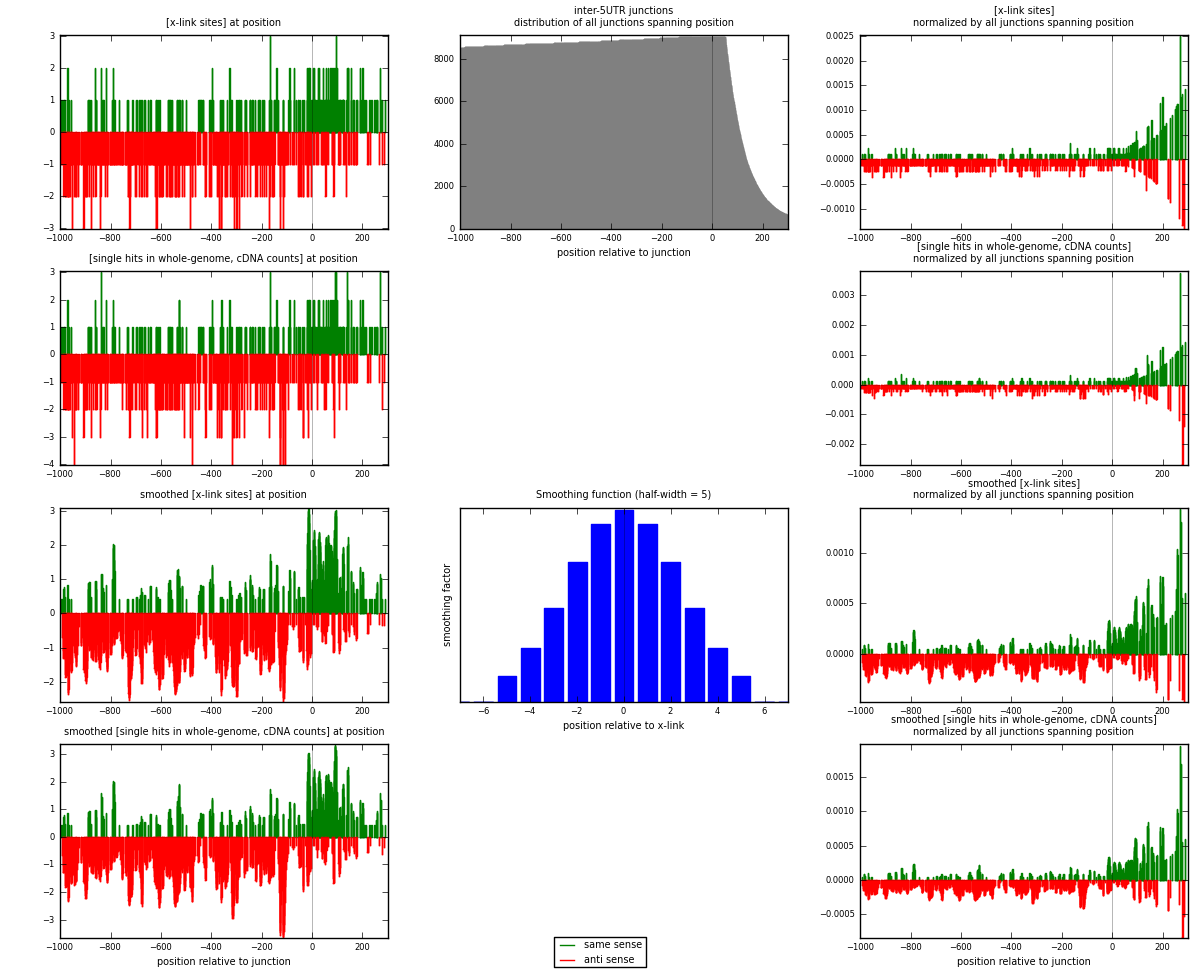 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 262 | 0.10% | 0.33% | 28.70% | 194 | 0.435216 |
| anti | 651 | 0.25% | 0.82% | 71.30% | 422 | 1.0814 |
| both | 913 | 0.35% | 1.15% | 100.00% | 602 | 1.51661 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 269 | 0.09% | 0.30% | 27.56% | 194 | 0.446844 |
| anti | 707 | 0.25% | 0.80% | 72.44% | 422 | 1.17442 |
| both | 976 | 0.34% | 1.10% | 100.00% | 602 | 1.62126 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
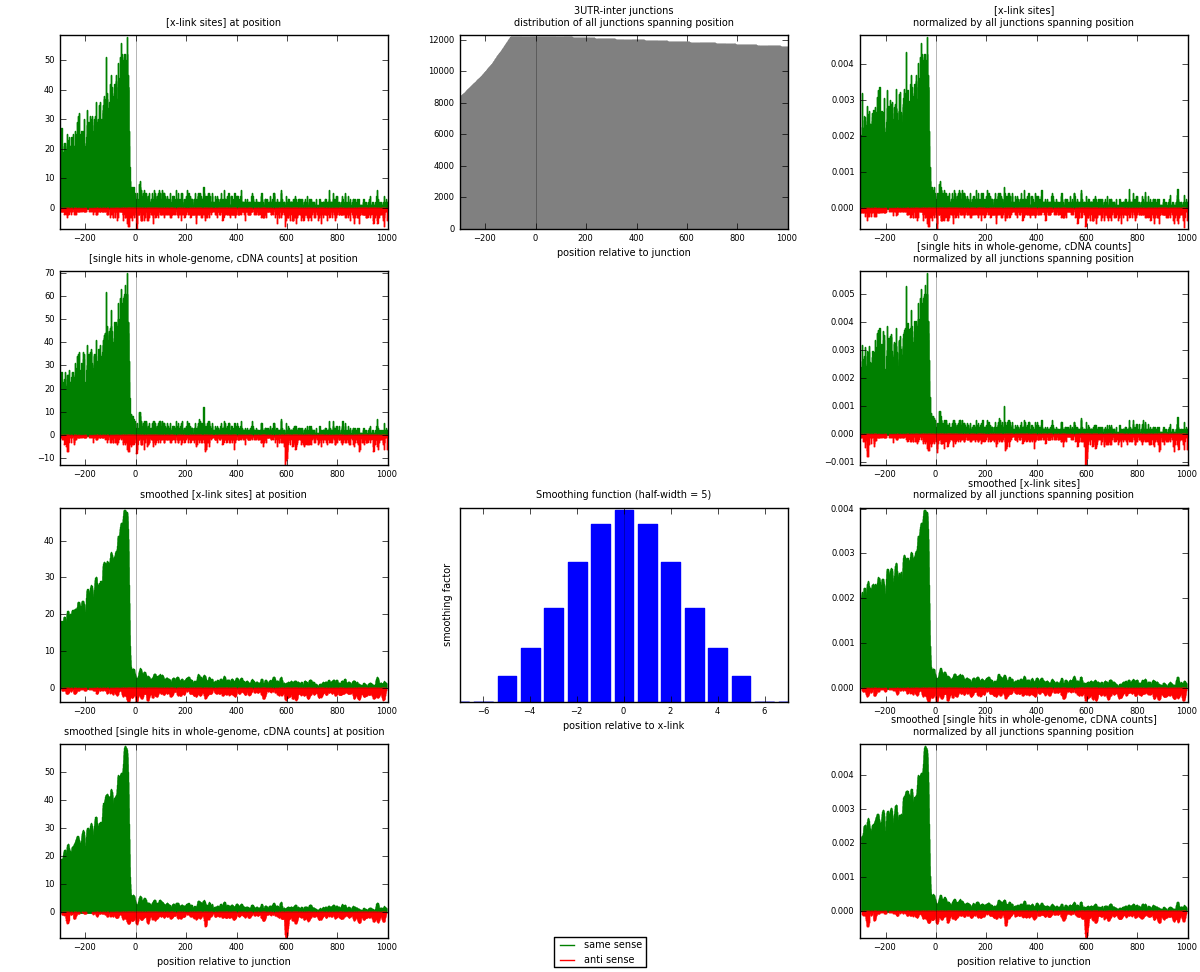 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9610 | 3.65% | 12.11% | 86.68% | 3223 | 2.63794 |
| anti | 1477 | 0.56% | 1.86% | 13.32% | 566 | 0.405435 |
| both | 11087 | 4.21% | 13.97% | 100.00% | 3643 | 3.04337 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 10840 | 3.77% | 12.27% | 86.29% | 3223 | 2.97557 |
| anti | 1722 | 0.60% | 1.95% | 13.71% | 566 | 0.472687 |
| both | 12562 | 4.37% | 14.21% | 100.00% | 3643 | 3.44826 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
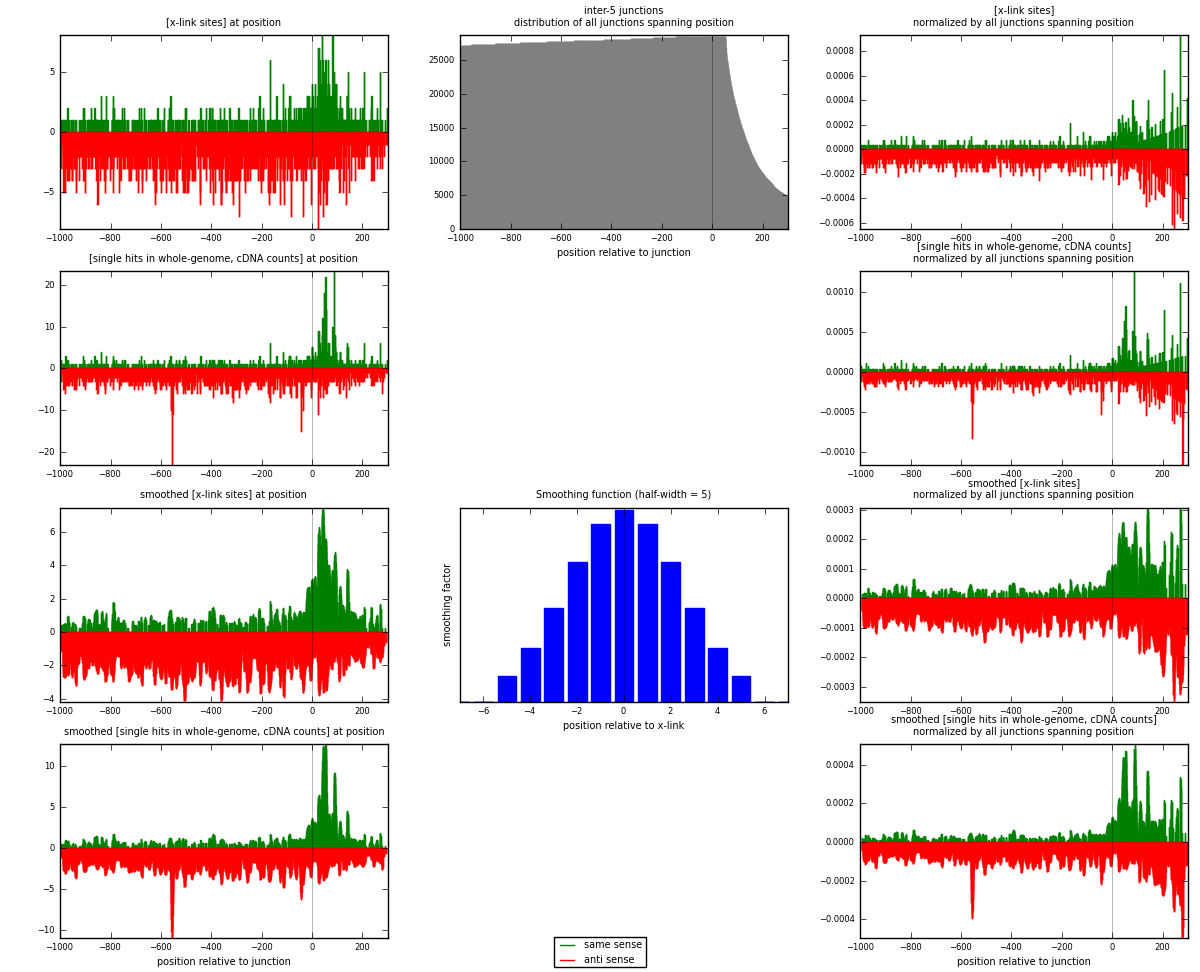
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 811 | 0.31% | 1.02% | 24.85% | 501 | 0.45613 |
| anti | 2452 | 0.93% | 3.09% | 75.15% | 1313 | 1.37908 |
| both | 3263 | 1.24% | 4.11% | 100.00% | 1778 | 1.83521 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 961 | 0.33% | 1.09% | 25.89% | 501 | 0.540495 |
| anti | 2751 | 0.96% | 3.11% | 74.11% | 1313 | 1.54724 |
| both | 3712 | 1.29% | 4.20% | 100.00% | 1778 | 2.08774 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
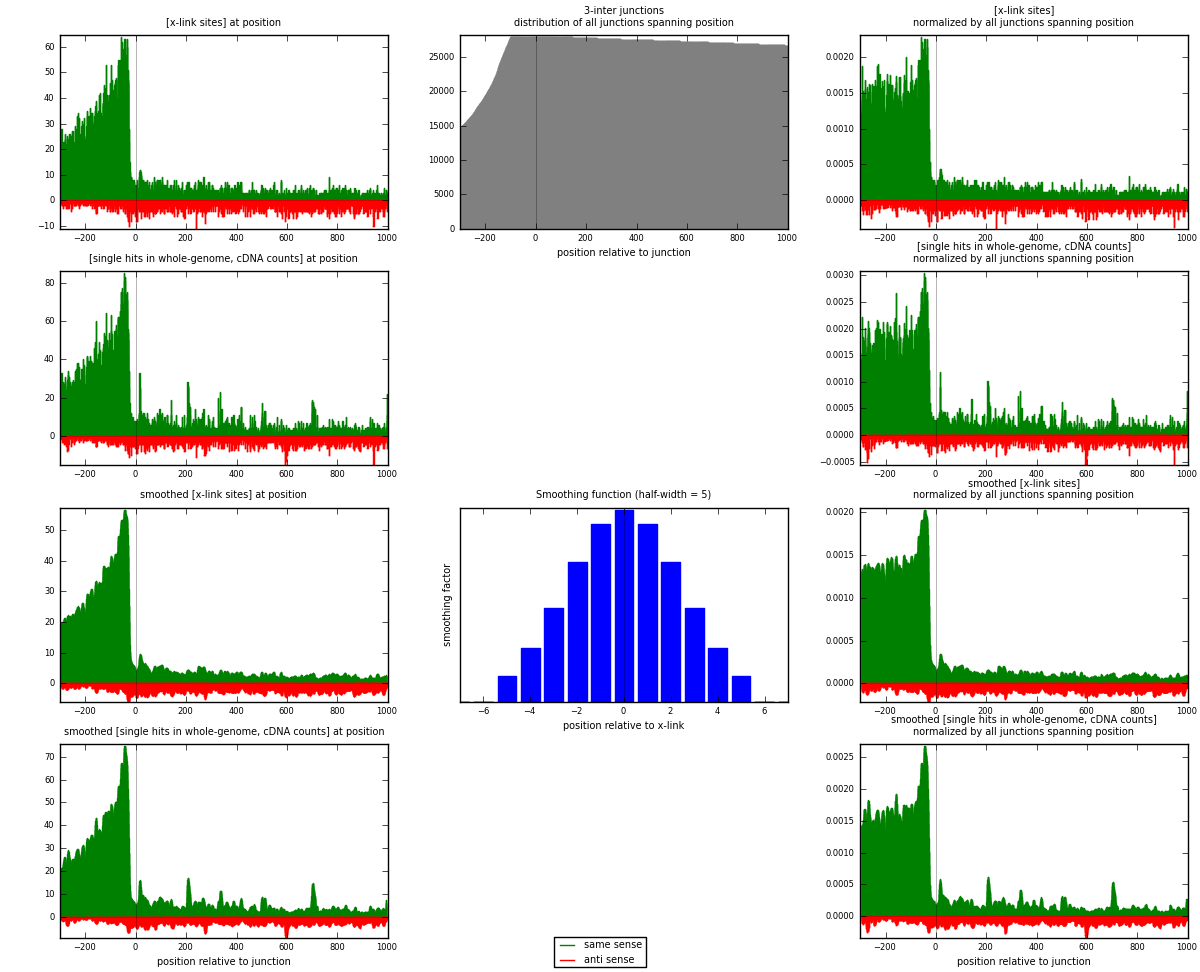
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 11941 | 4.53% | 15.05% | 80.15% | 3980 | 2.38629 |
| anti | 2957 | 1.12% | 3.73% | 19.85% | 1210 | 0.590927 |
| both | 14898 | 5.66% | 18.78% | 100.00% | 5004 | 2.97722 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 14911 | 5.19% | 16.87% | 81.57% | 3980 | 2.97982 |
| anti | 3369 | 1.17% | 3.81% | 18.43% | 1210 | 0.673261 |
| both | 18280 | 6.36% | 20.68% | 100.00% | 5004 | 3.65308 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
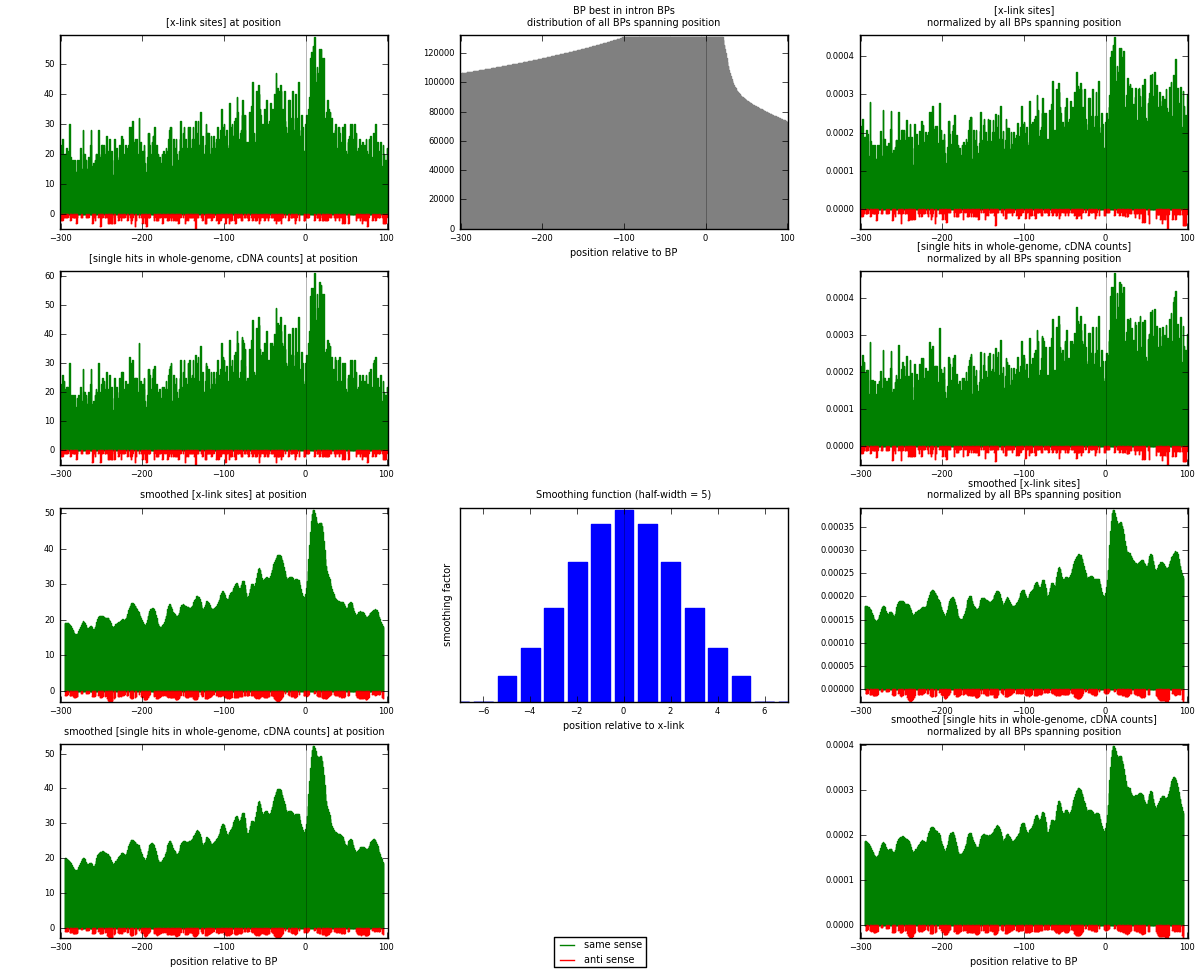
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 10193 | 3.87% | 12.85% | 96.31% | 8153 | 1.20485 |
| anti | 391 | 0.15% | 0.49% | 3.69% | 321 | 0.0462175 |
| both | 10584 | 4.02% | 13.34% | 100.00% | 8460 | 1.25106 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 10611 | 3.69% | 12.01% | 96.33% | 8153 | 1.25426 |
| anti | 404 | 0.14% | 0.46% | 3.67% | 321 | 0.0477541 |
| both | 11015 | 3.83% | 12.46% | 100.00% | 8460 | 1.30201 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.