RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
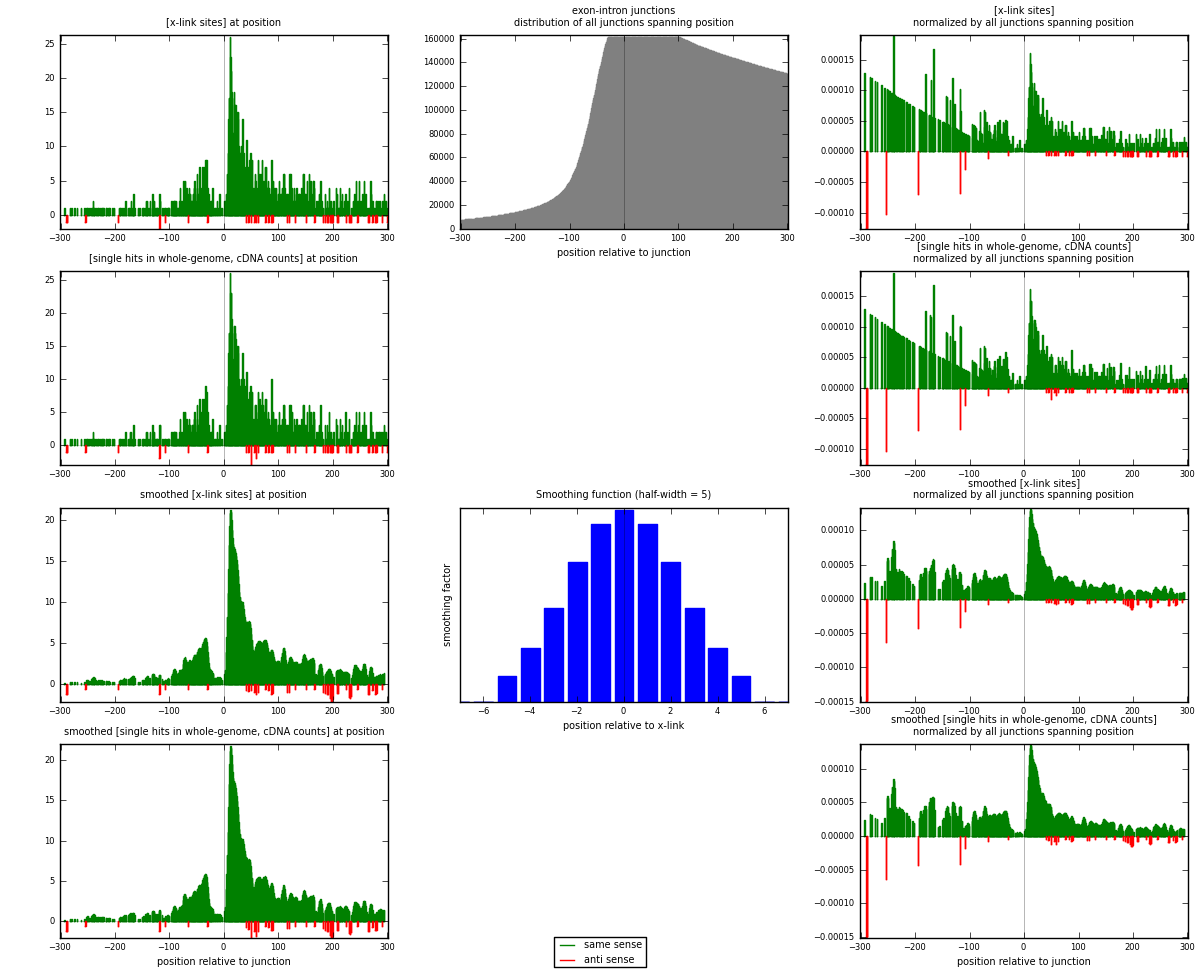 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1389 | 8.55% | 25.39% | 96.79% | 1294 | 1.04436 |
| anti | 46 | 0.28% | 0.84% | 3.21% | 36 | 0.0345865 |
| both | 1435 | 8.83% | 26.23% | 100.00% | 1330 | 1.07895 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1428 | 8.32% | 24.69% | 96.68% | 1294 | 1.07368 |
| anti | 49 | 0.29% | 0.85% | 3.32% | 36 | 0.0368421 |
| both | 1477 | 8.60% | 25.54% | 100.00% | 1330 | 1.11053 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
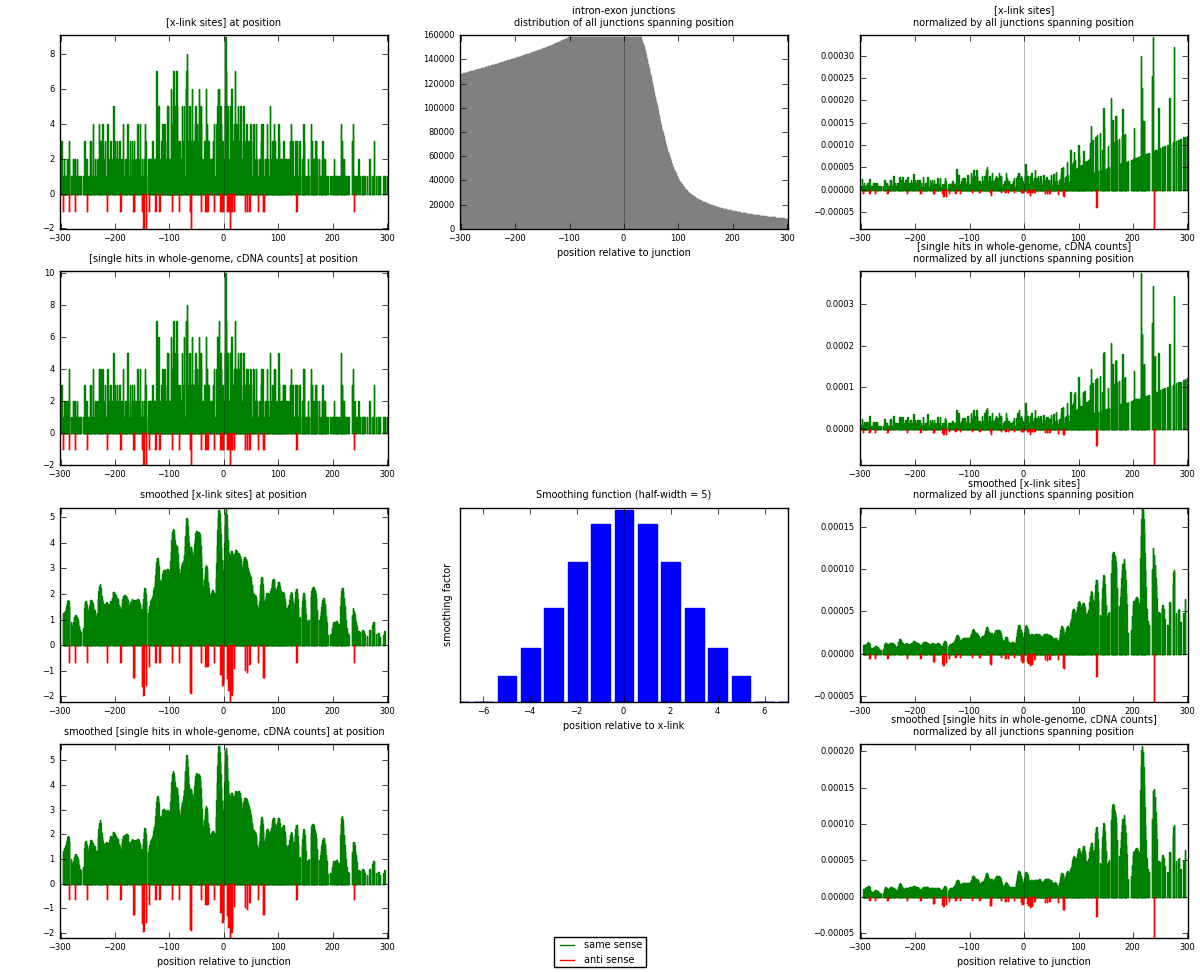 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1054 | 6.49% | 19.27% | 96.08% | 957 | 1.05717 |
| anti | 43 | 0.26% | 0.79% | 3.92% | 40 | 0.0431294 |
| both | 1097 | 6.75% | 20.05% | 100.00% | 997 | 1.1003 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1084 | 6.31% | 18.74% | 96.18% | 957 | 1.08726 |
| anti | 43 | 0.25% | 0.74% | 3.82% | 40 | 0.0431294 |
| both | 1127 | 6.56% | 19.48% | 100.00% | 997 | 1.13039 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
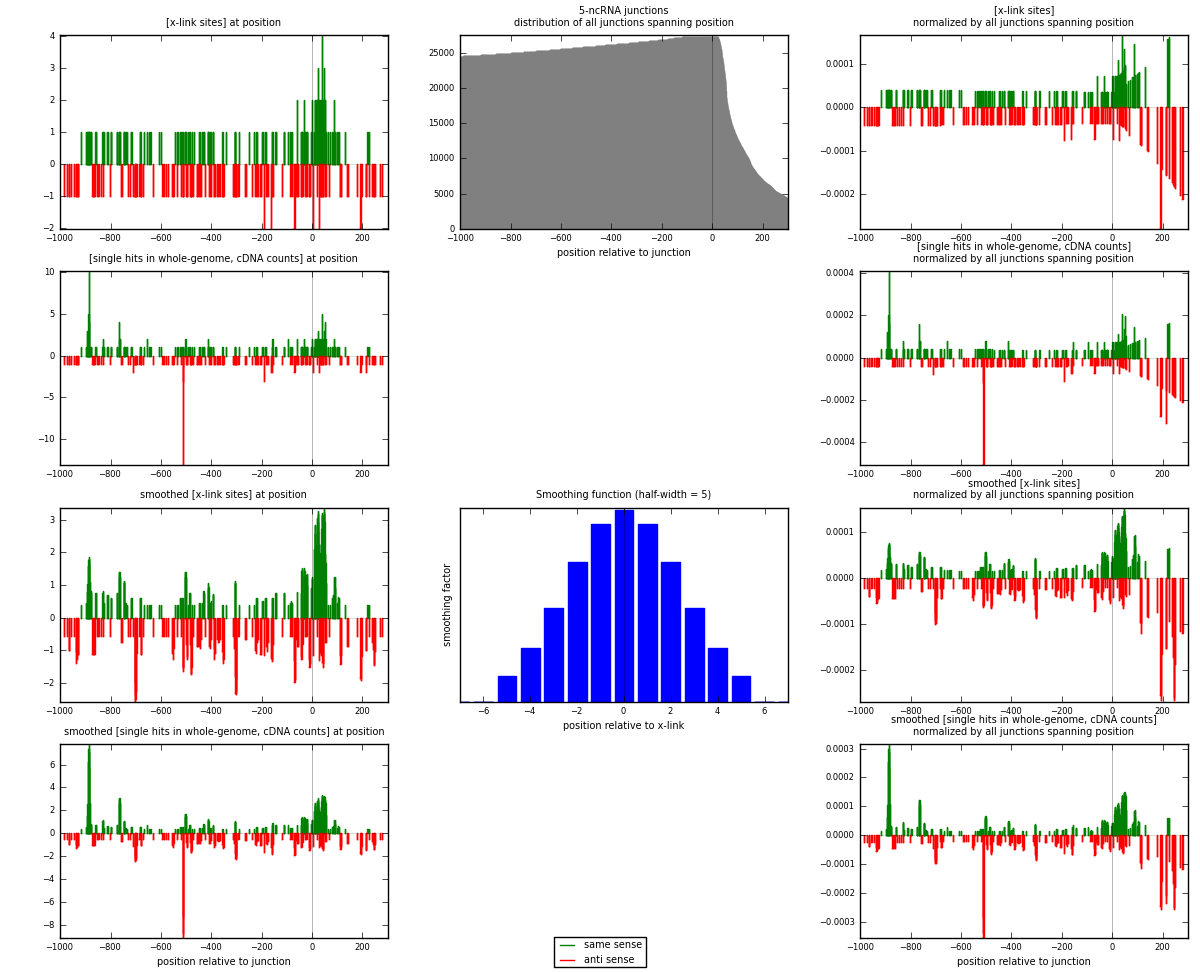 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 177 | 1.09% | 3.24% | 53.64% | 88 | 0.898477 |
| anti | 153 | 0.94% | 2.80% | 46.36% | 110 | 0.77665 |
| both | 330 | 2.03% | 6.03% | 100.00% | 197 | 1.67513 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 211 | 1.23% | 3.65% | 55.24% | 88 | 1.07107 |
| anti | 171 | 1.00% | 2.96% | 44.76% | 110 | 0.86802 |
| both | 382 | 2.22% | 6.60% | 100.00% | 197 | 1.93909 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
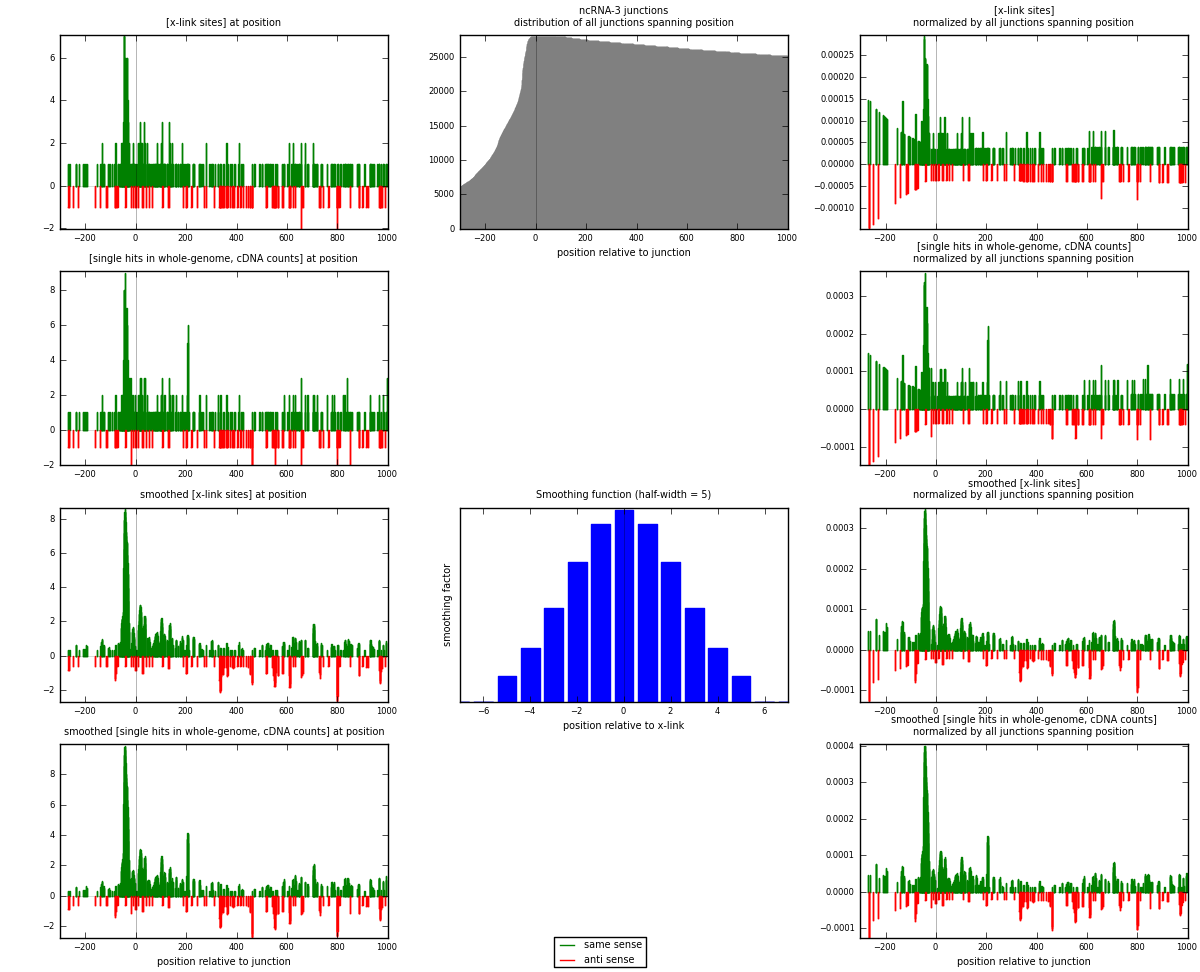 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 381 | 2.34% | 6.97% | 78.23% | 211 | 1.31834 |
| anti | 106 | 0.65% | 1.94% | 21.77% | 78 | 0.366782 |
| both | 487 | 3.00% | 8.90% | 100.00% | 289 | 1.68512 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 434 | 2.53% | 7.50% | 79.63% | 211 | 1.50173 |
| anti | 111 | 0.65% | 1.92% | 20.37% | 78 | 0.384083 |
| both | 545 | 3.17% | 9.42% | 100.00% | 289 | 1.88581 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
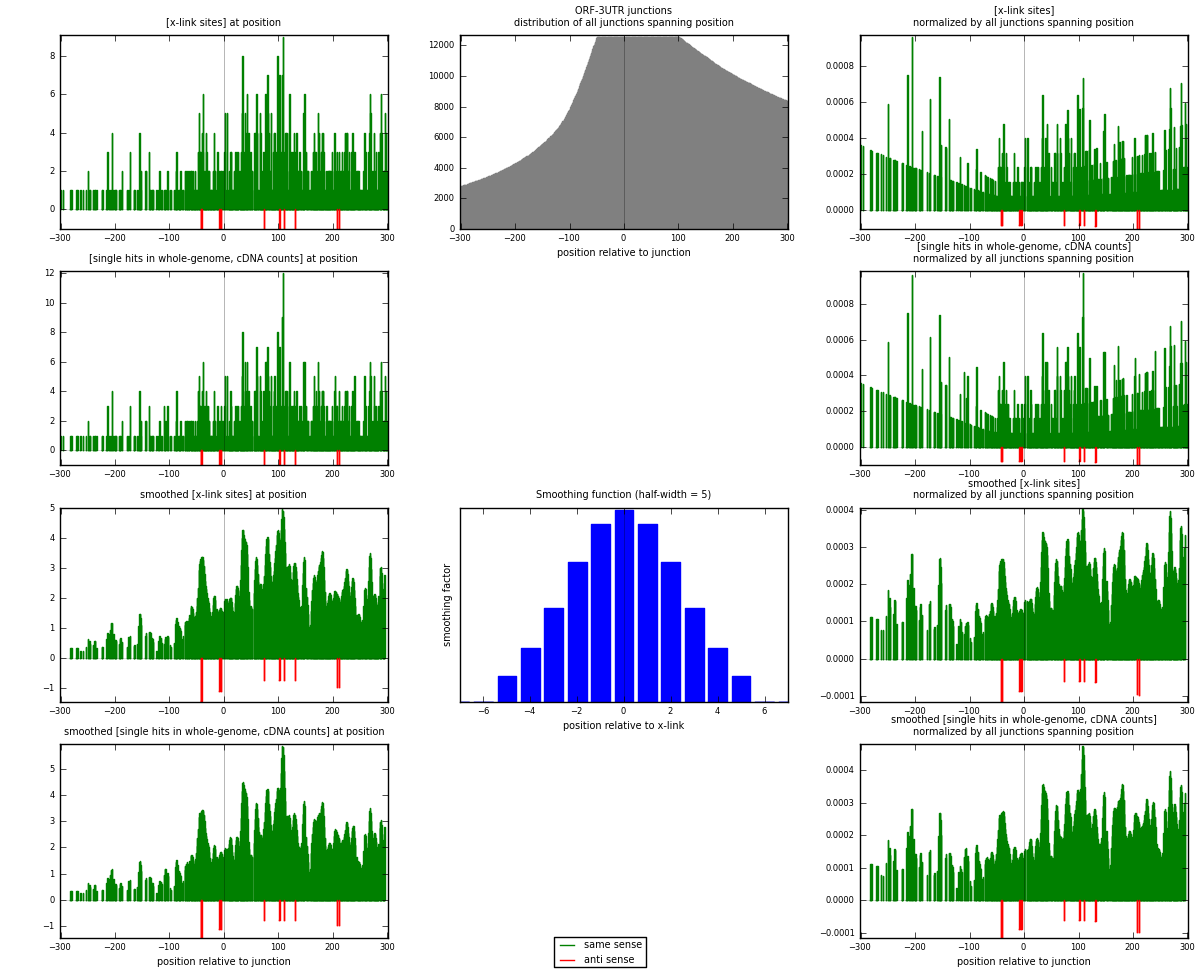 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 854 | 5.26% | 15.61% | 98.84% | 555 | 1.5115 |
| anti | 10 | 0.06% | 0.18% | 1.16% | 10 | 0.0176991 |
| both | 864 | 5.32% | 15.80% | 100.00% | 565 | 1.5292 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 904 | 5.26% | 15.63% | 98.91% | 555 | 1.6 |
| anti | 10 | 0.06% | 0.17% | 1.09% | 10 | 0.0176991 |
| both | 914 | 5.32% | 15.80% | 100.00% | 565 | 1.6177 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
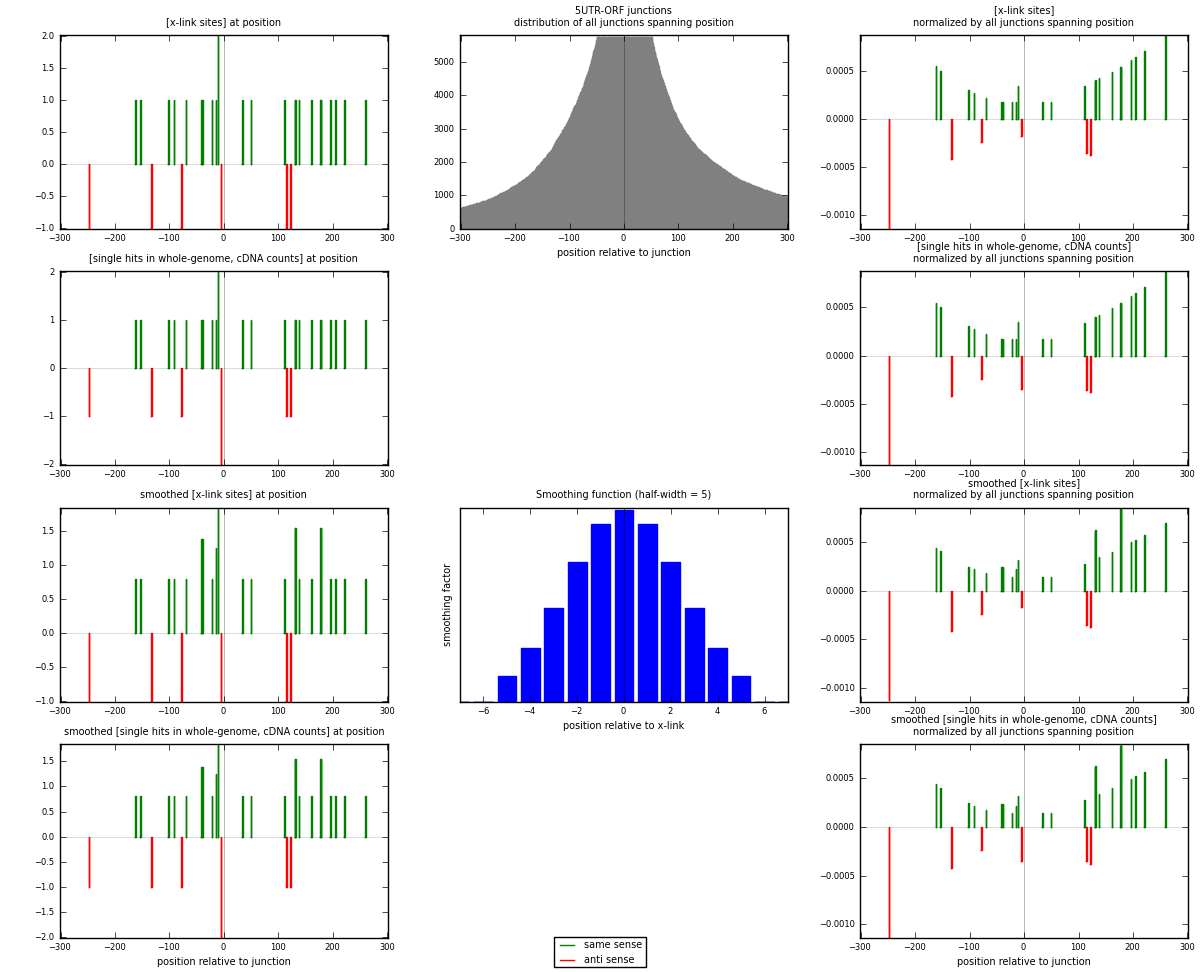 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 24 | 0.15% | 0.44% | 80.00% | 23 | 0.857143 |
| anti | 6 | 0.04% | 0.11% | 20.00% | 5 | 0.214286 |
| both | 30 | 0.18% | 0.55% | 100.00% | 28 | 1.07143 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 24 | 0.14% | 0.41% | 77.42% | 23 | 0.857143 |
| anti | 7 | 0.04% | 0.12% | 22.58% | 5 | 0.25 |
| both | 31 | 0.18% | 0.54% | 100.00% | 28 | 1.10714 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
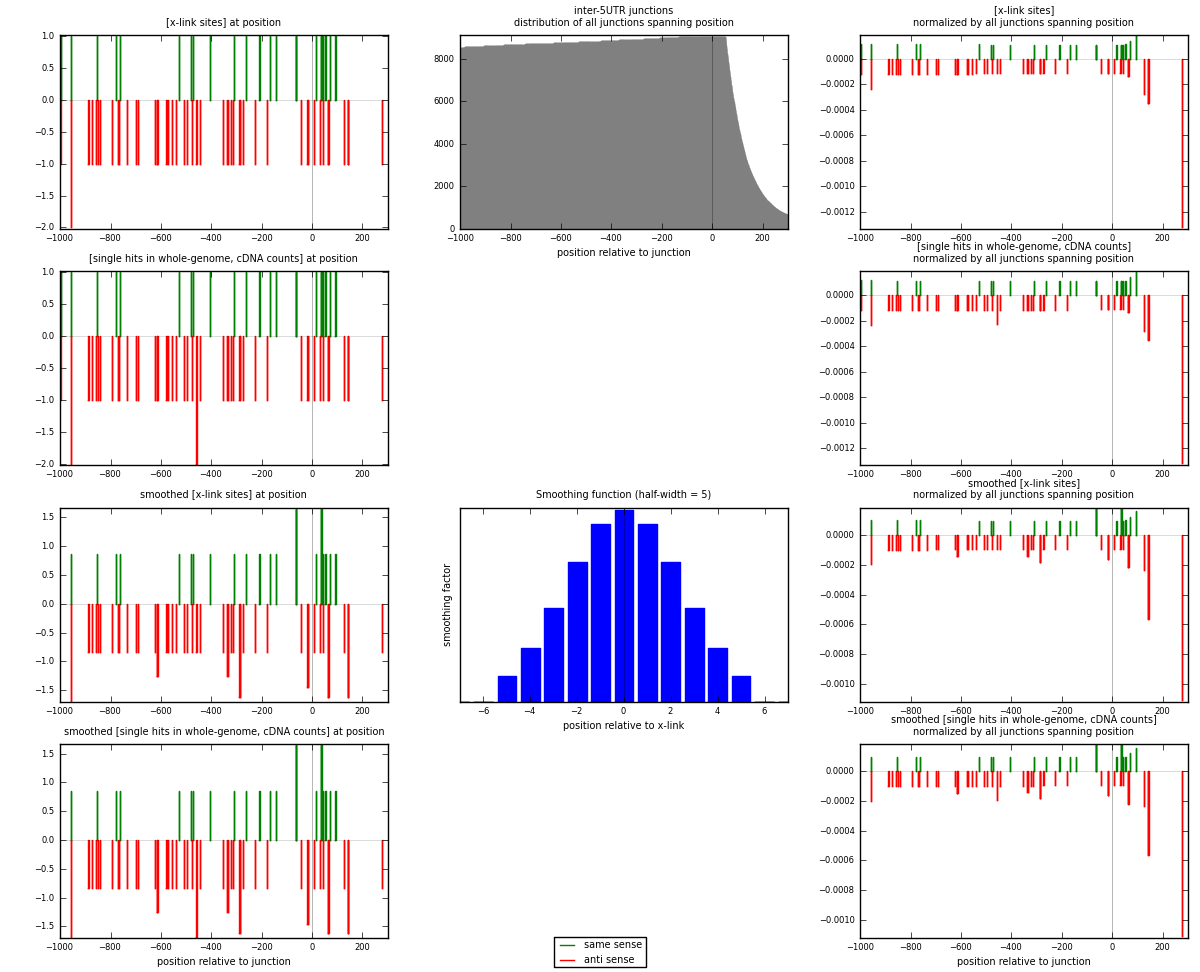 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 23 | 0.14% | 0.42% | 32.39% | 21 | 0.370968 |
| anti | 48 | 0.30% | 0.88% | 67.61% | 41 | 0.774194 |
| both | 71 | 0.44% | 1.30% | 100.00% | 62 | 1.14516 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 23 | 0.13% | 0.40% | 31.94% | 21 | 0.370968 |
| anti | 49 | 0.29% | 0.85% | 68.06% | 41 | 0.790323 |
| both | 72 | 0.42% | 1.24% | 100.00% | 62 | 1.16129 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
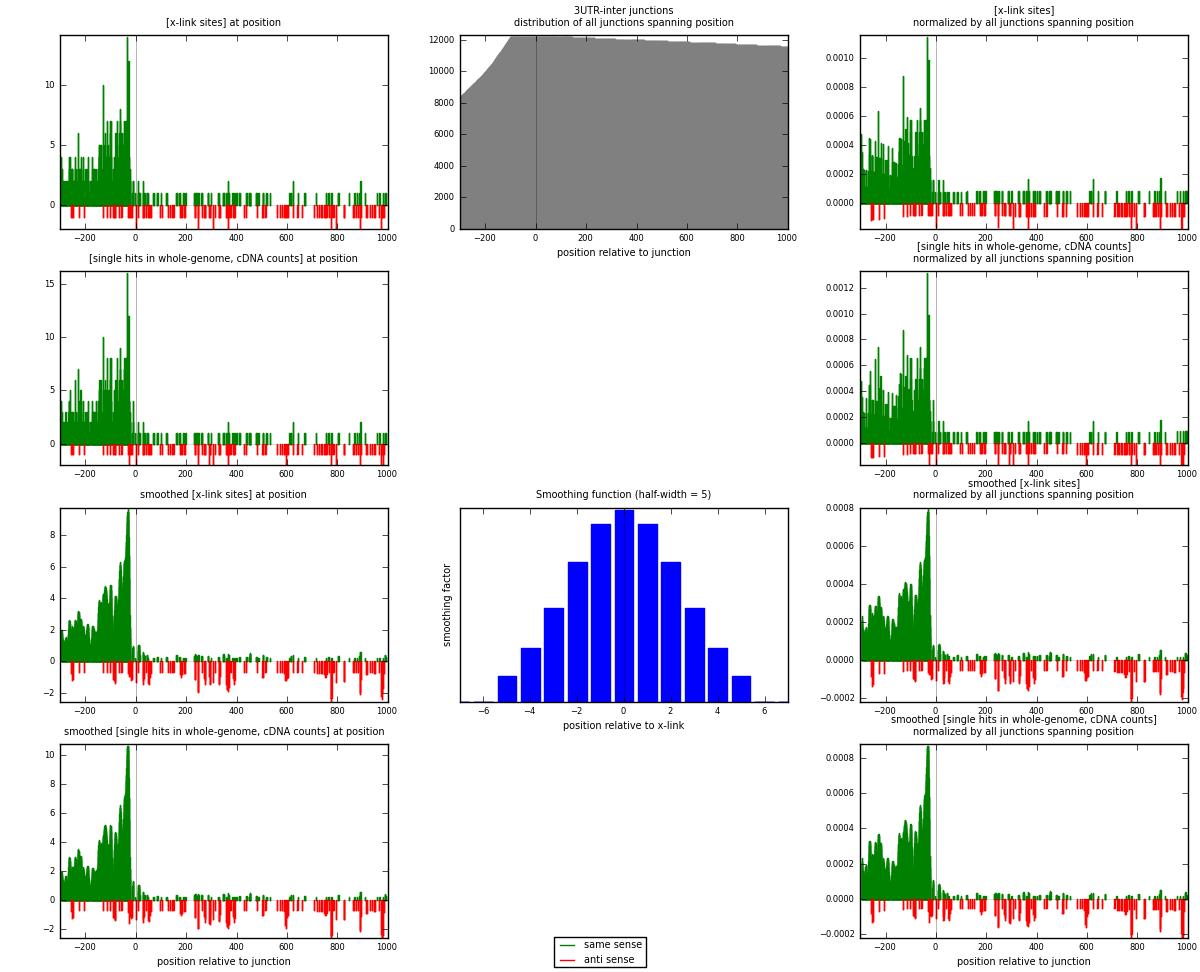 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 851 | 5.24% | 15.56% | 87.19% | 595 | 1.25702 |
| anti | 125 | 0.77% | 2.29% | 12.81% | 87 | 0.184638 |
| both | 976 | 6.01% | 17.84% | 100.00% | 677 | 1.44165 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 901 | 5.25% | 15.58% | 87.48% | 595 | 1.33087 |
| anti | 129 | 0.75% | 2.23% | 12.52% | 87 | 0.190547 |
| both | 1030 | 6.00% | 17.81% | 100.00% | 677 | 1.52142 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
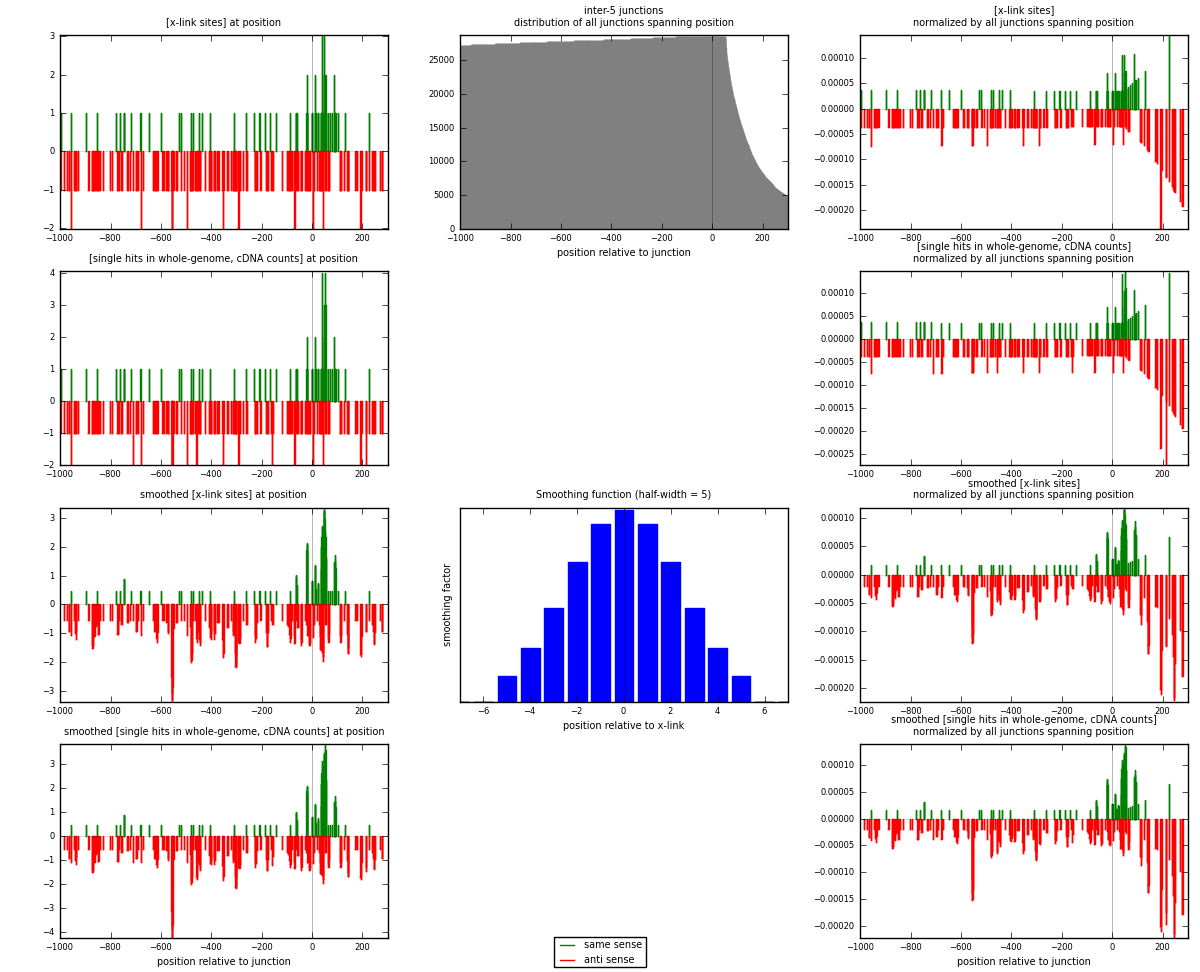
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 75 | 0.46% | 1.37% | 28.41% | 58 | 0.388601 |
| anti | 189 | 1.16% | 3.46% | 71.59% | 136 | 0.979275 |
| both | 264 | 1.62% | 4.83% | 100.00% | 193 | 1.36788 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 79 | 0.46% | 1.37% | 28.83% | 58 | 0.409326 |
| anti | 195 | 1.14% | 3.37% | 71.17% | 136 | 1.01036 |
| both | 274 | 1.60% | 4.74% | 100.00% | 193 | 1.41969 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
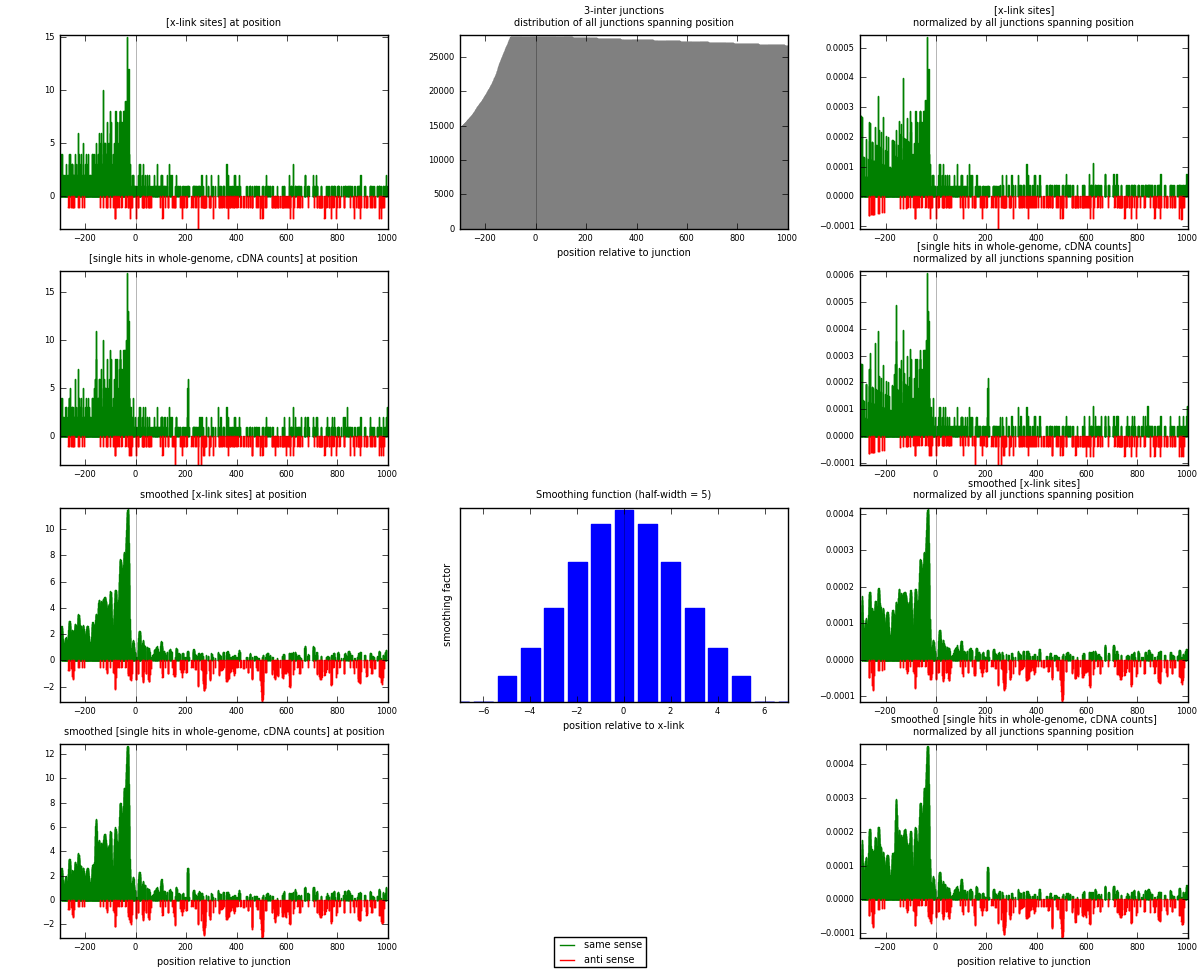
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1168 | 7.19% | 21.35% | 83.01% | 739 | 1.32426 |
| anti | 239 | 1.47% | 4.37% | 16.99% | 149 | 0.270975 |
| both | 1407 | 8.66% | 25.72% | 100.00% | 882 | 1.59524 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1281 | 7.46% | 22.15% | 83.45% | 739 | 1.45238 |
| anti | 254 | 1.48% | 4.39% | 16.55% | 149 | 0.287982 |
| both | 1535 | 8.94% | 26.54% | 100.00% | 882 | 1.74036 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
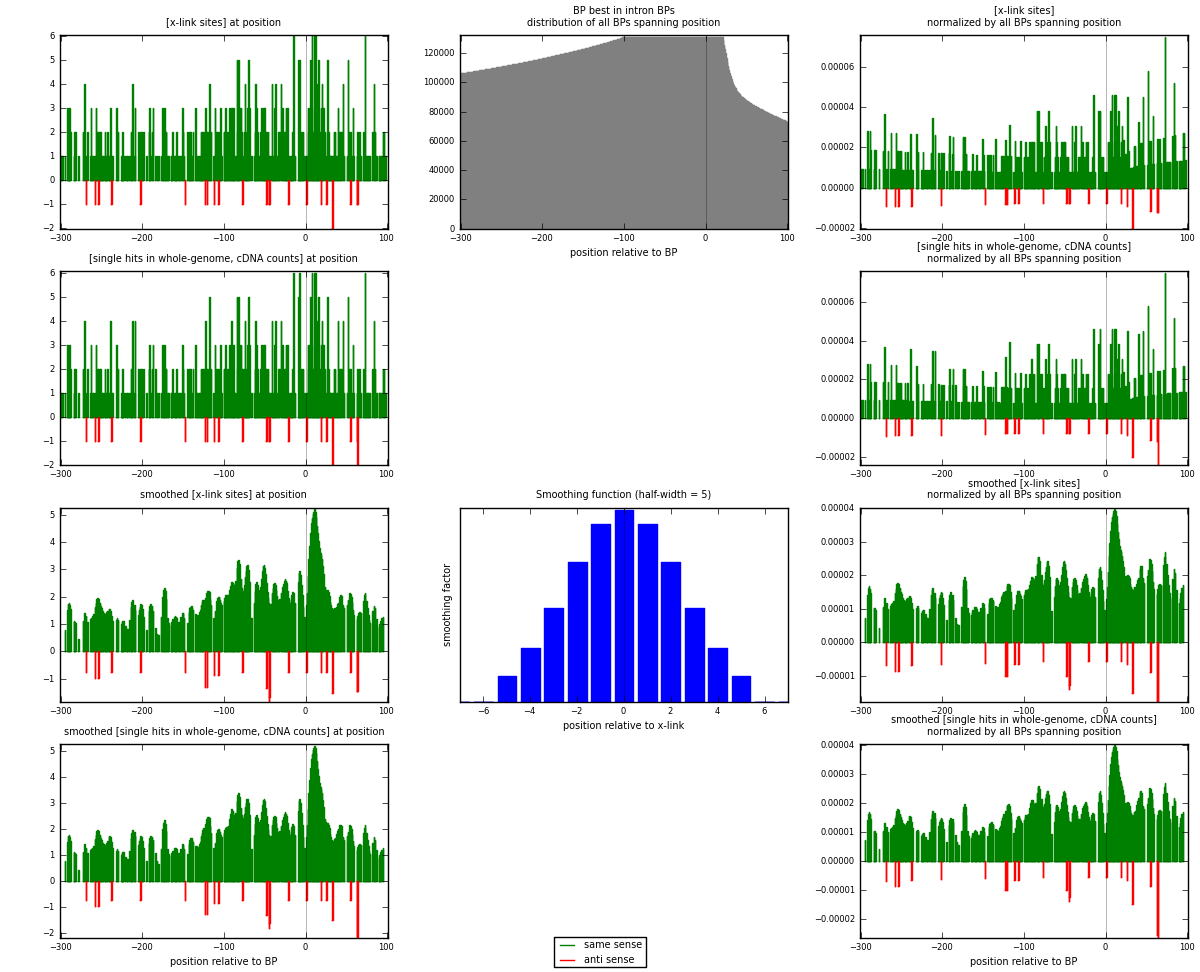
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 547 | 3.37% | 10.00% | 95.96% | 511 | 1.02434 |
| anti | 23 | 0.14% | 0.42% | 4.04% | 23 | 0.0430712 |
| both | 570 | 3.51% | 10.42% | 100.00% | 534 | 1.06742 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 556 | 3.24% | 9.61% | 95.86% | 511 | 1.0412 |
| anti | 24 | 0.14% | 0.41% | 4.14% | 23 | 0.0449438 |
| both | 580 | 3.38% | 10.03% | 100.00% | 534 | 1.08614 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.