RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
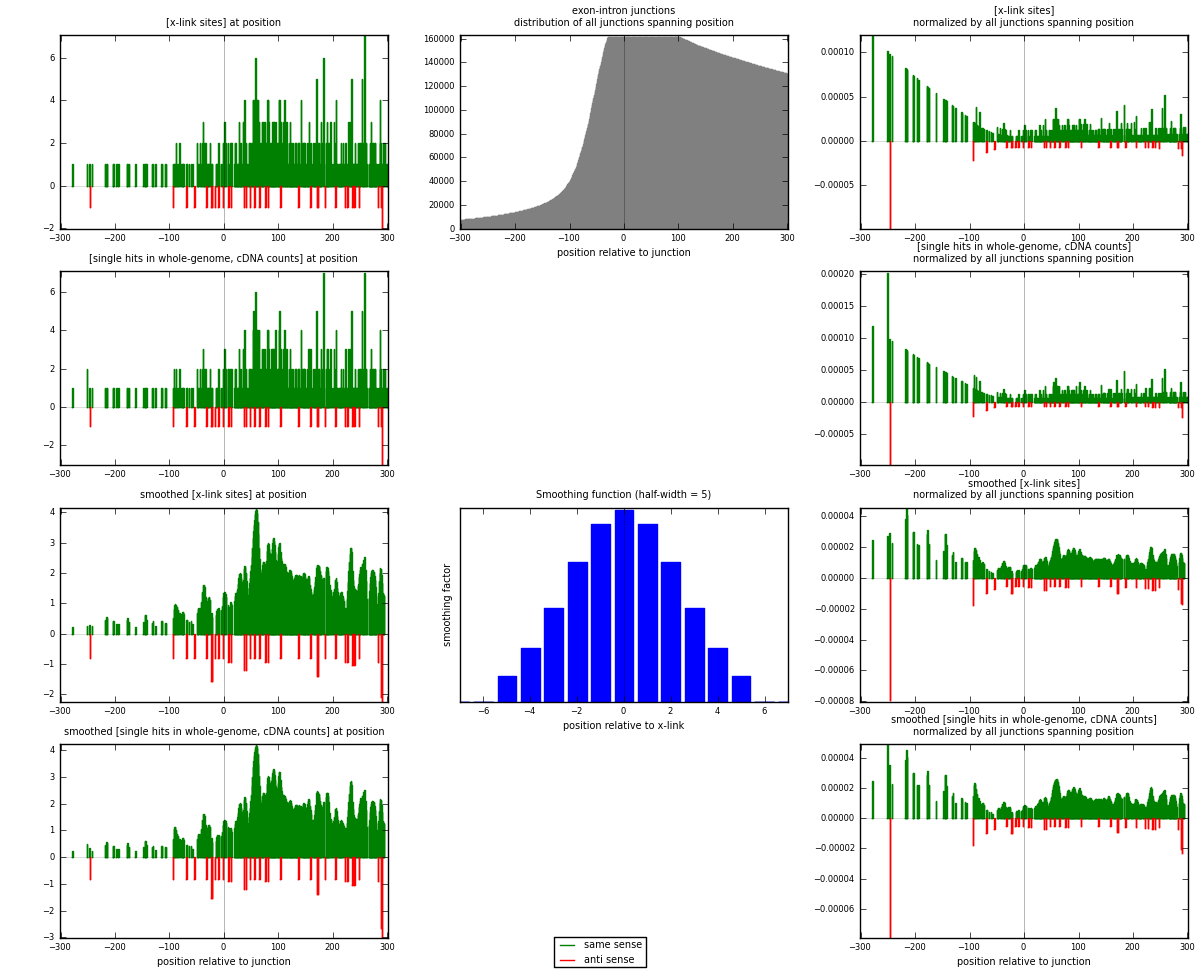 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 526 | 3.98% | 27.35% | 93.76% | 475 | 1.03137 |
| anti | 35 | 0.26% | 1.82% | 6.24% | 35 | 0.0686275 |
| both | 561 | 4.25% | 29.17% | 100.00% | 510 | 1.1 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 540 | 3.92% | 24.94% | 93.75% | 475 | 1.05882 |
| anti | 36 | 0.26% | 1.66% | 6.25% | 35 | 0.0705882 |
| both | 576 | 4.18% | 26.61% | 100.00% | 510 | 1.12941 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
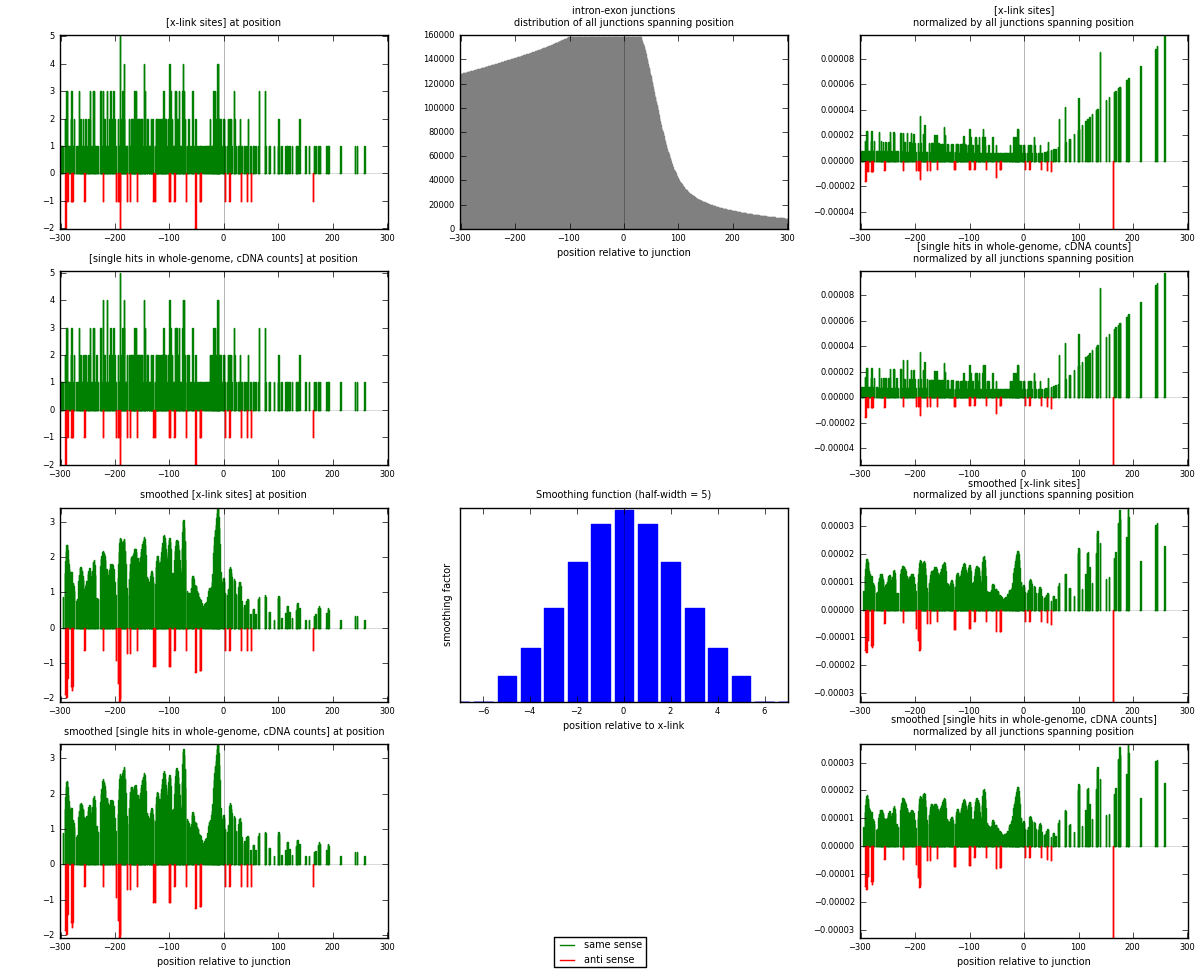 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 427 | 3.23% | 22.20% | 92.83% | 395 | 1 |
| anti | 33 | 0.25% | 1.72% | 7.17% | 32 | 0.0772834 |
| both | 460 | 3.48% | 23.92% | 100.00% | 427 | 1.07728 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 438 | 3.18% | 20.23% | 92.99% | 395 | 1.02576 |
| anti | 33 | 0.24% | 1.52% | 7.01% | 32 | 0.0772834 |
| both | 471 | 3.42% | 21.76% | 100.00% | 427 | 1.10304 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
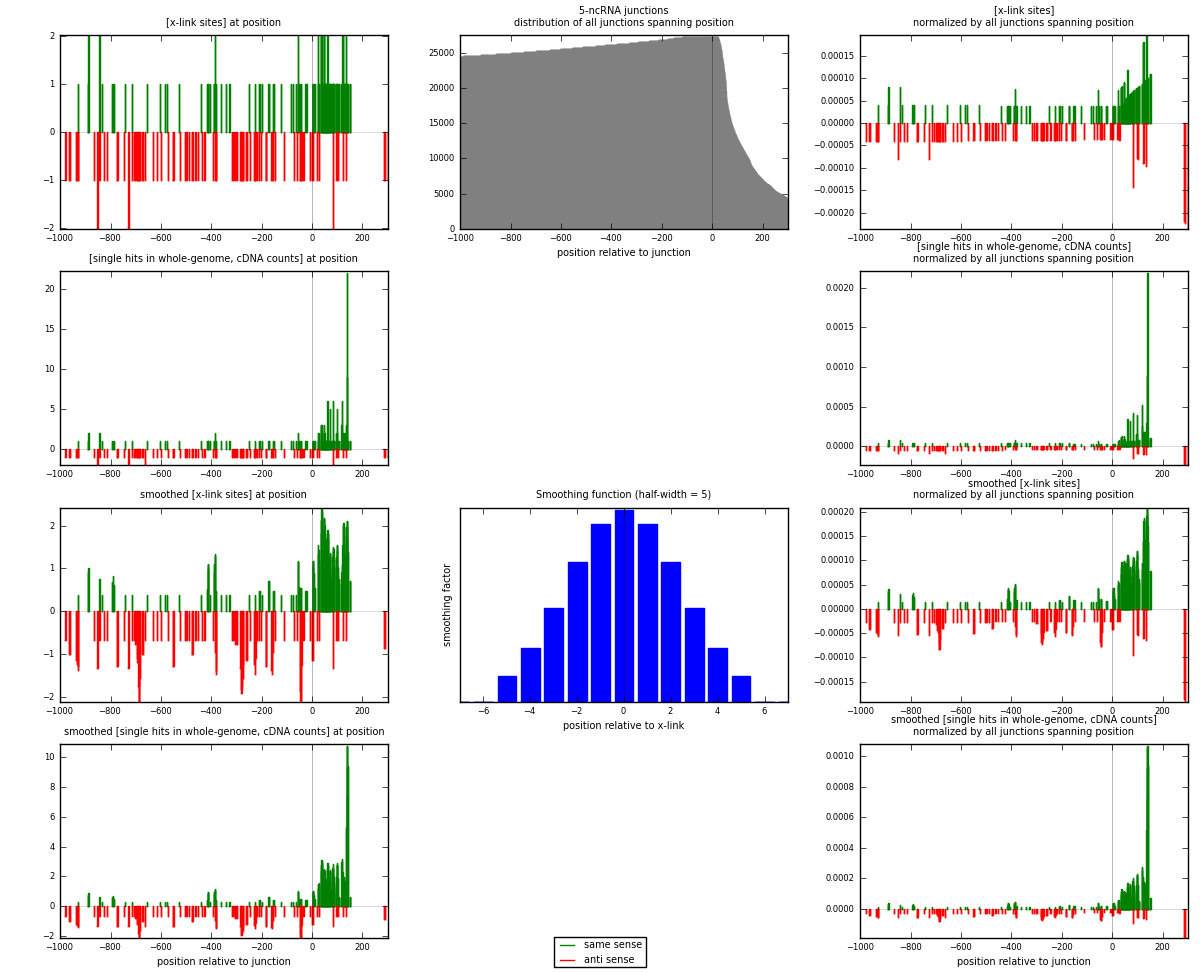 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 131 | 0.99% | 6.81% | 57.46% | 56 | 0.916084 |
| anti | 97 | 0.73% | 5.04% | 42.54% | 87 | 0.678322 |
| both | 228 | 1.73% | 11.86% | 100.00% | 143 | 1.59441 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 197 | 1.43% | 9.10% | 66.78% | 56 | 1.37762 |
| anti | 98 | 0.71% | 4.53% | 33.22% | 87 | 0.685315 |
| both | 295 | 2.14% | 13.63% | 100.00% | 143 | 2.06294 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
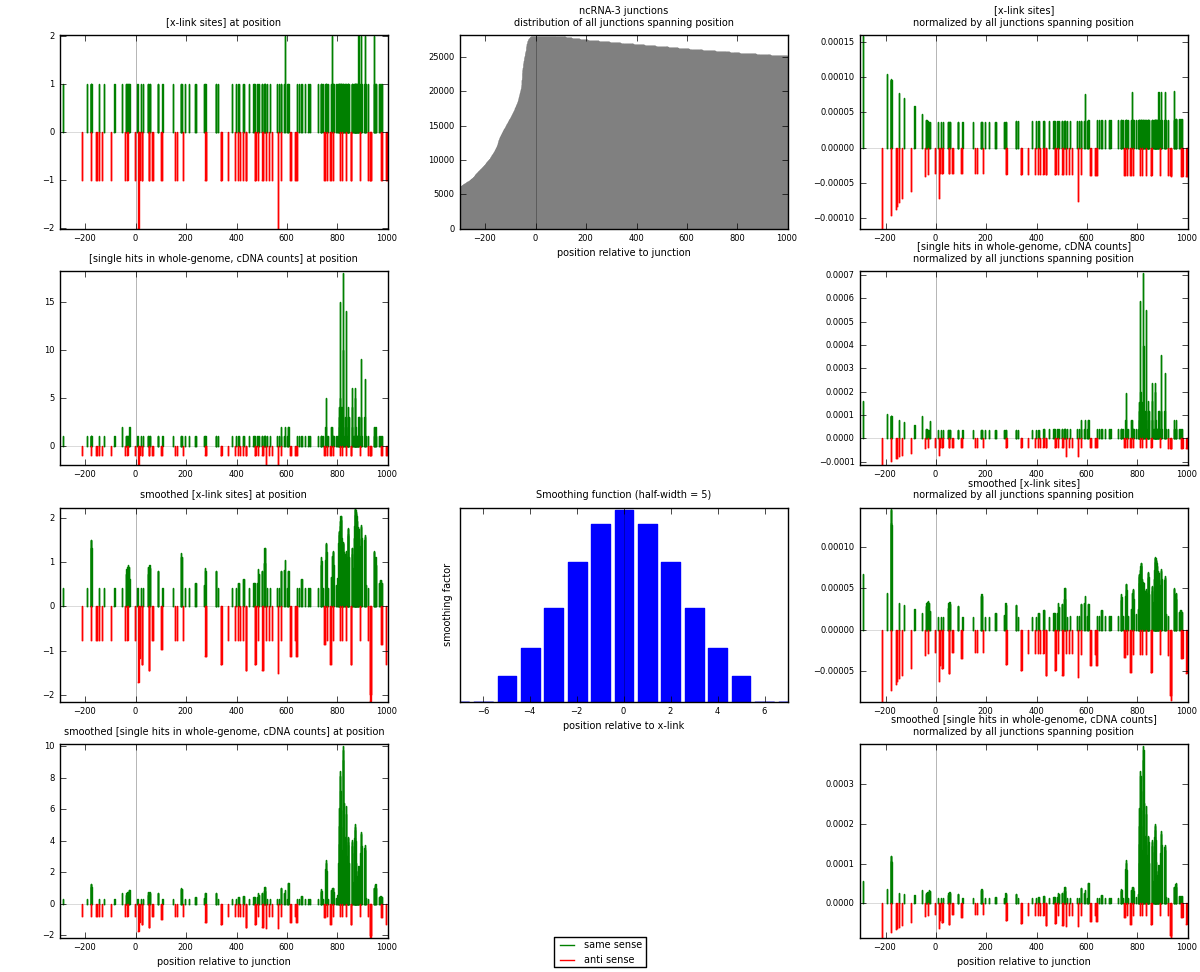 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 161 | 1.22% | 8.37% | 69.40% | 58 | 1.33058 |
| anti | 71 | 0.54% | 3.69% | 30.60% | 63 | 0.586777 |
| both | 232 | 1.76% | 12.06% | 100.00% | 121 | 1.91736 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 293 | 2.13% | 13.53% | 80.27% | 58 | 2.42149 |
| anti | 72 | 0.52% | 3.33% | 19.73% | 63 | 0.595041 |
| both | 365 | 2.65% | 16.86% | 100.00% | 121 | 3.01653 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
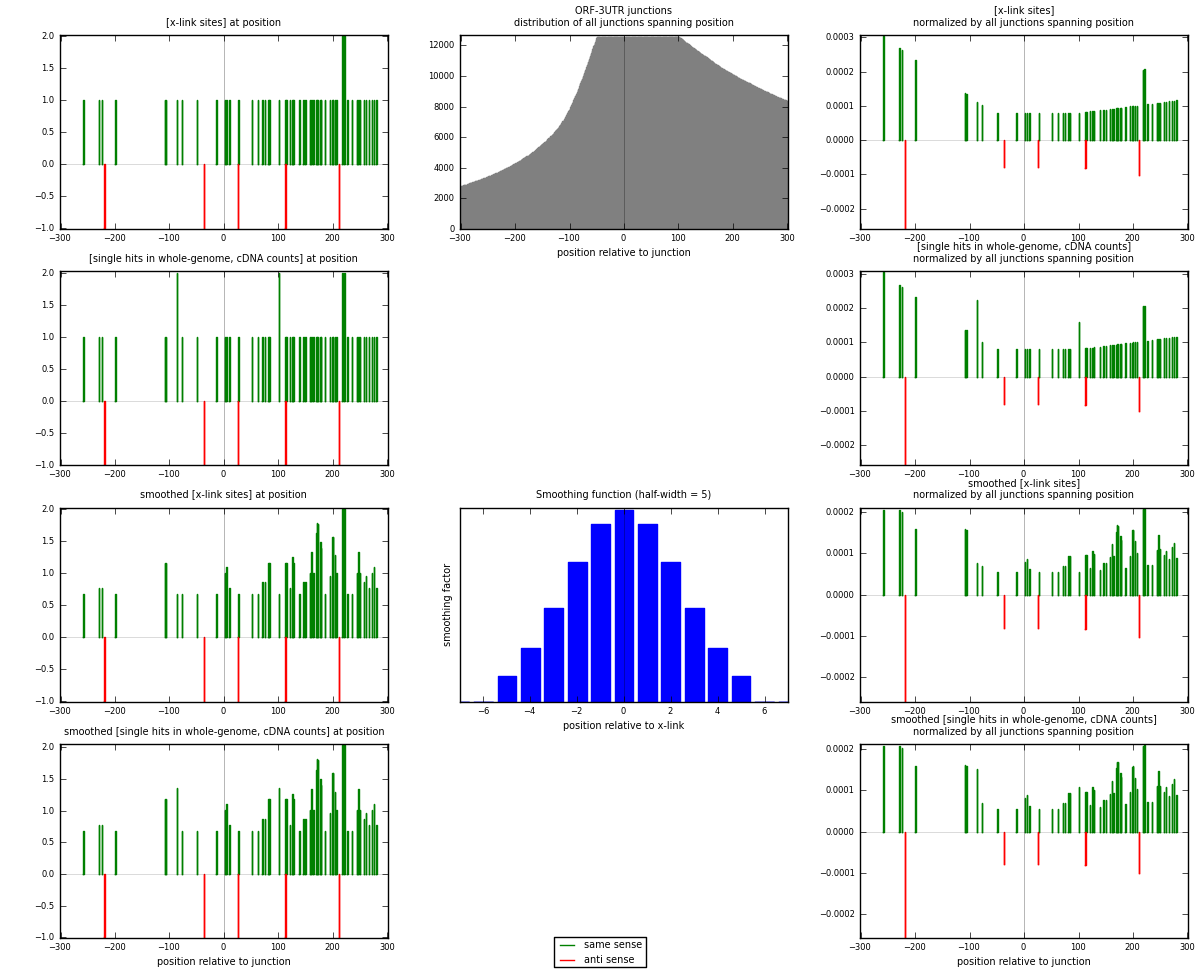 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 58 | 0.44% | 3.02% | 92.06% | 54 | 0.983051 |
| anti | 5 | 0.04% | 0.26% | 7.94% | 5 | 0.0847458 |
| both | 63 | 0.48% | 3.28% | 100.00% | 59 | 1.0678 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 60 | 0.44% | 2.77% | 92.31% | 54 | 1.01695 |
| anti | 5 | 0.04% | 0.23% | 7.69% | 5 | 0.0847458 |
| both | 65 | 0.47% | 3.00% | 100.00% | 59 | 1.10169 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
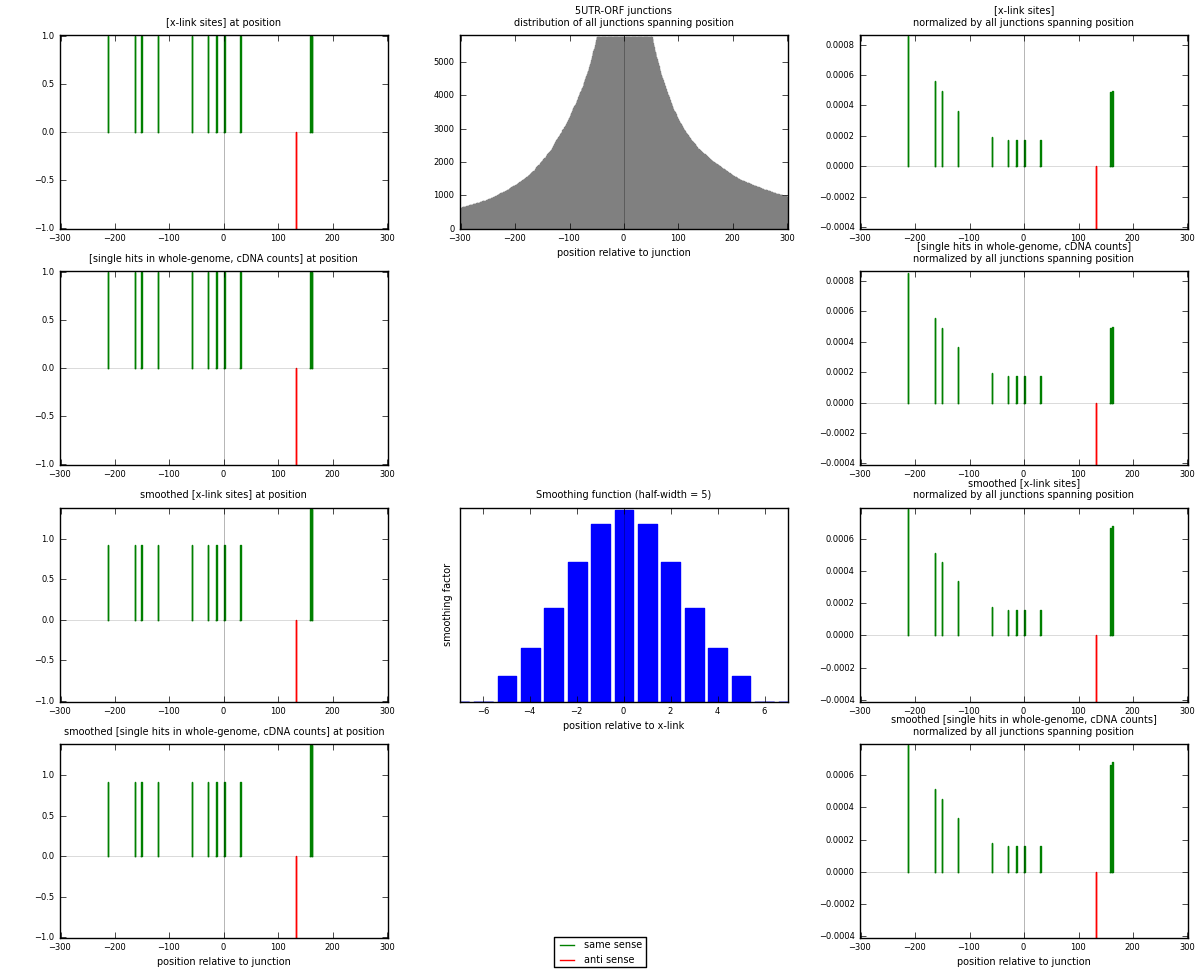 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 11 | 0.08% | 0.57% | 91.67% | 11 | 0.916667 |
| anti | 1 | 0.01% | 0.05% | 8.33% | 1 | 0.0833333 |
| both | 12 | 0.09% | 0.62% | 100.00% | 12 | 1 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 11 | 0.08% | 0.51% | 91.67% | 11 | 0.916667 |
| anti | 1 | 0.01% | 0.05% | 8.33% | 1 | 0.0833333 |
| both | 12 | 0.09% | 0.55% | 100.00% | 12 | 1 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
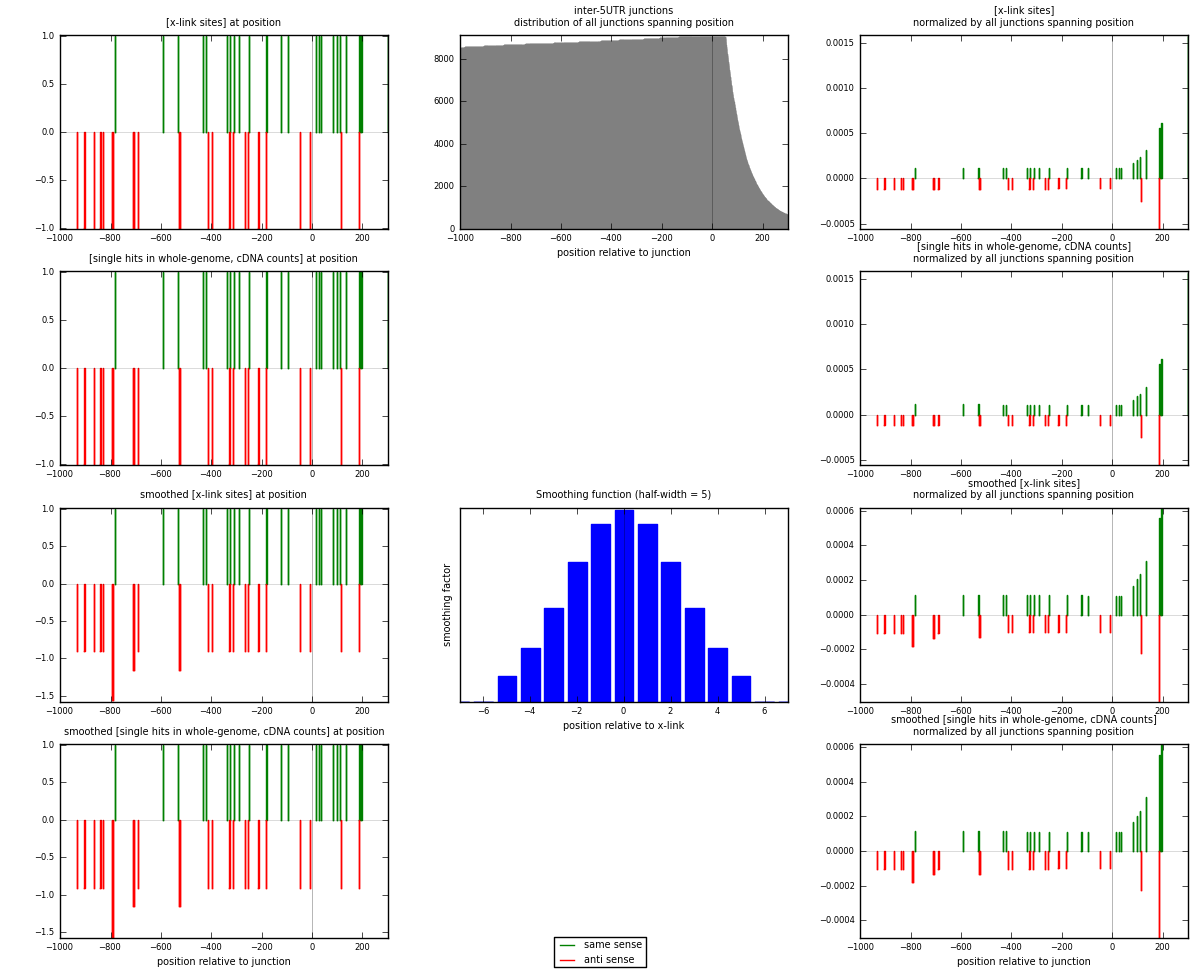 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 23 | 0.17% | 1.20% | 48.94% | 22 | 0.522727 |
| anti | 24 | 0.18% | 1.25% | 51.06% | 22 | 0.545455 |
| both | 47 | 0.36% | 2.44% | 100.00% | 44 | 1.06818 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 23 | 0.17% | 1.06% | 48.94% | 22 | 0.522727 |
| anti | 24 | 0.17% | 1.11% | 51.06% | 22 | 0.545455 |
| both | 47 | 0.34% | 2.17% | 100.00% | 44 | 1.06818 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
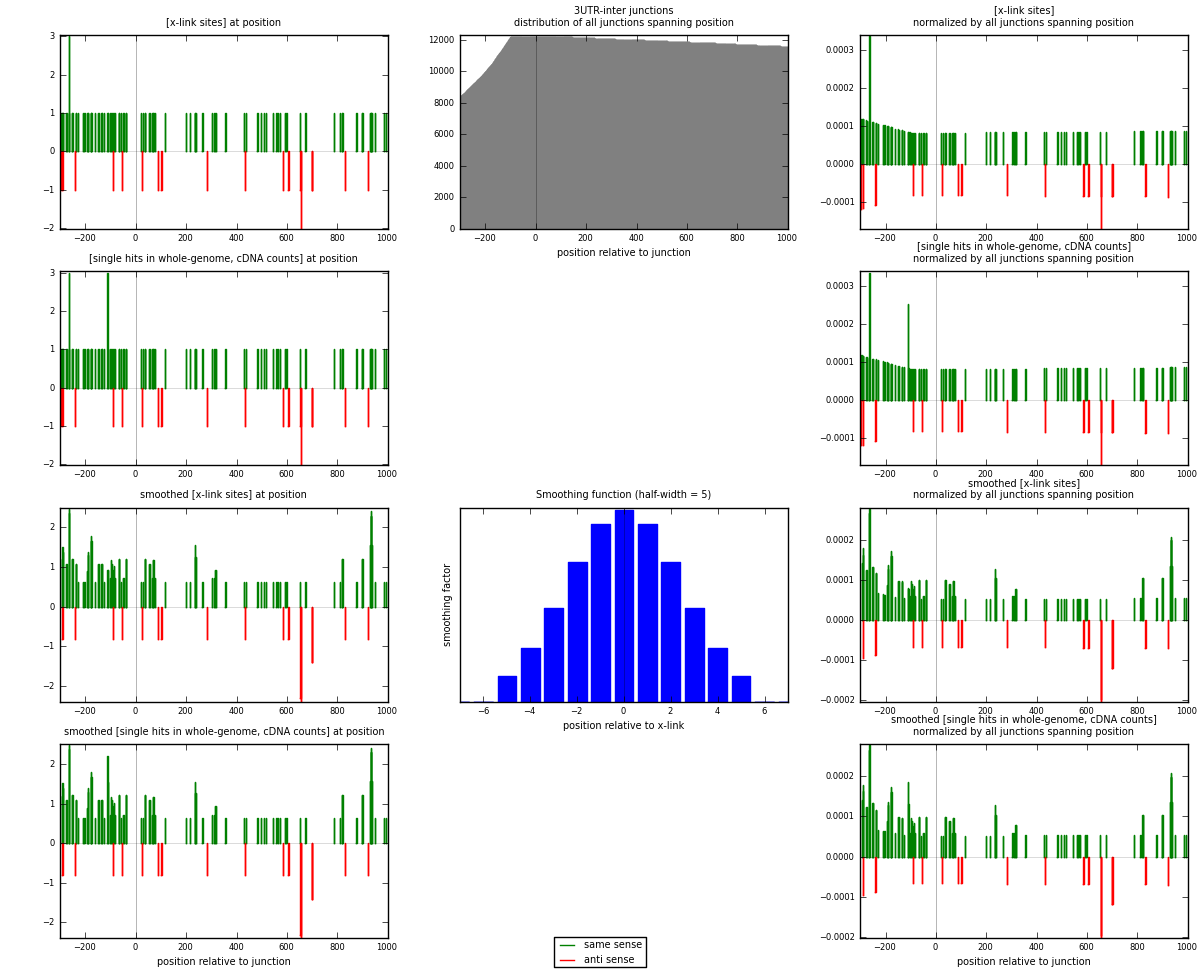 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 96 | 0.73% | 4.99% | 83.48% | 83 | 0.950495 |
| anti | 19 | 0.14% | 0.99% | 16.52% | 18 | 0.188119 |
| both | 115 | 0.87% | 5.98% | 100.00% | 101 | 1.13861 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 98 | 0.71% | 4.53% | 83.76% | 83 | 0.970297 |
| anti | 19 | 0.14% | 0.88% | 16.24% | 18 | 0.188119 |
| both | 117 | 0.85% | 5.40% | 100.00% | 101 | 1.15842 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
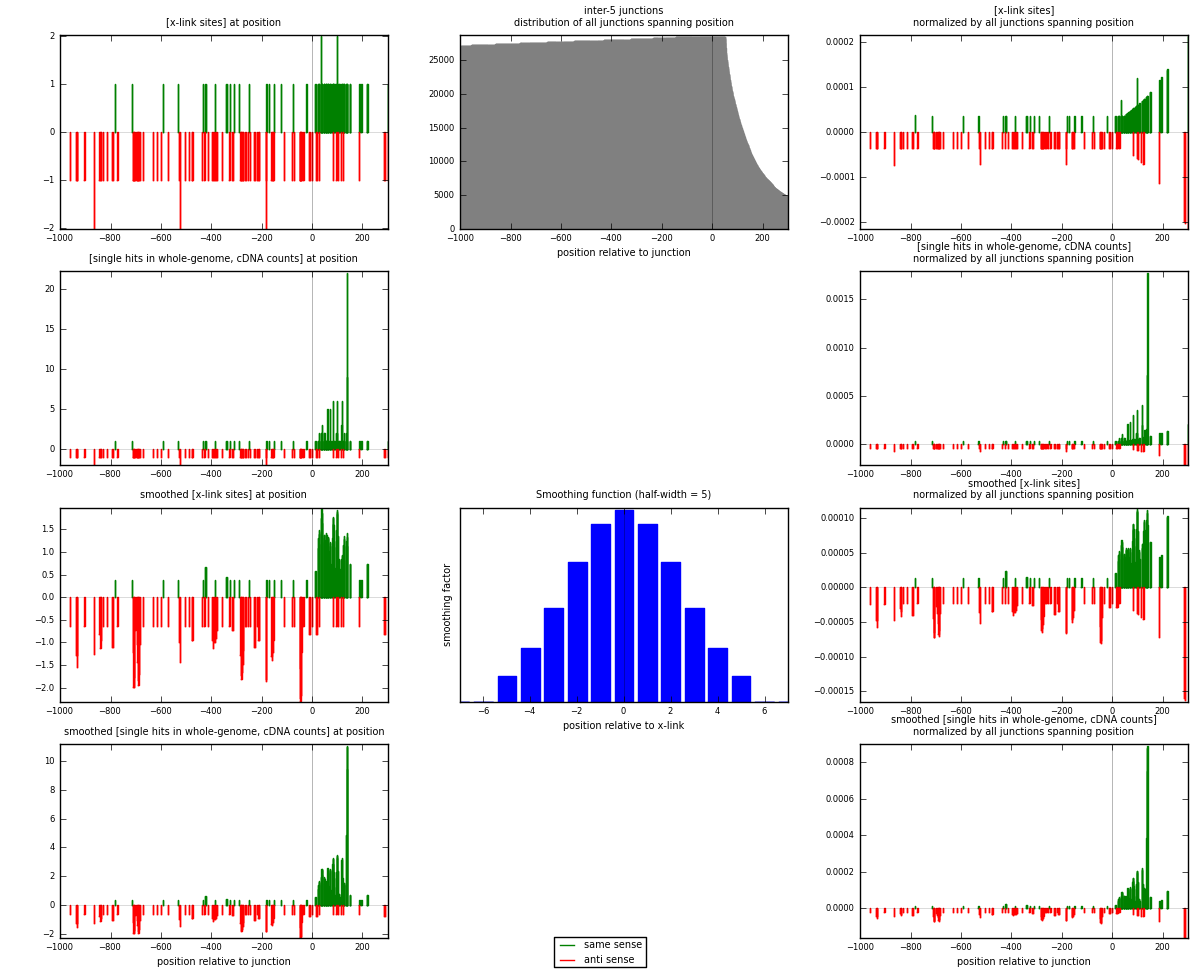 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 86 | 0.65% | 4.47% | 46.99% | 40 | 0.68254 |
| anti | 97 | 0.73% | 5.04% | 53.01% | 86 | 0.769841 |
| both | 183 | 1.38% | 9.52% | 100.00% | 126 | 1.45238 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 149 | 1.08% | 6.88% | 60.57% | 40 | 1.18254 |
| anti | 97 | 0.70% | 4.48% | 39.43% | 86 | 0.769841 |
| both | 246 | 1.78% | 11.36% | 100.00% | 126 | 1.95238 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
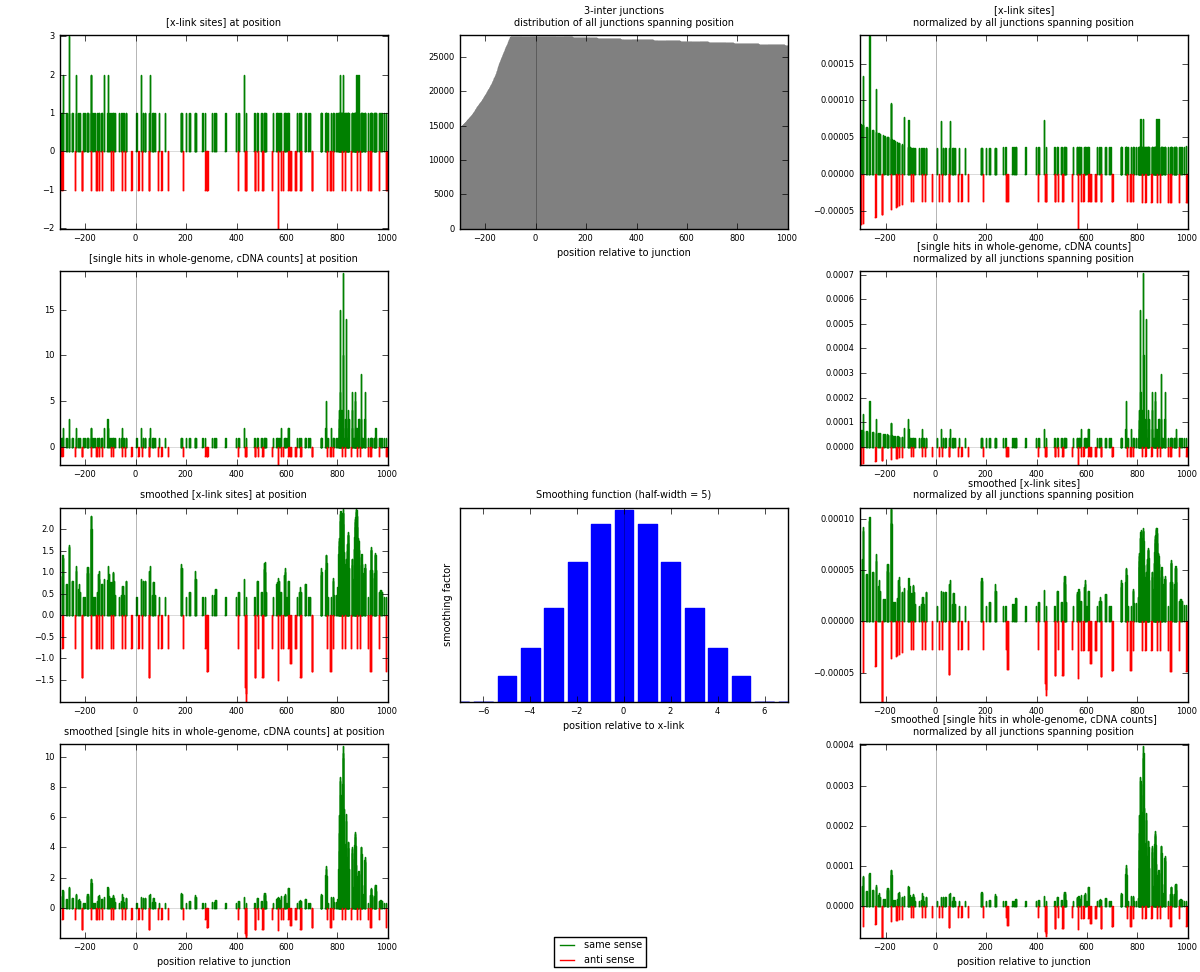 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 220 | 1.67% | 11.44% | 77.46% | 106 | 1.34969 |
| anti | 64 | 0.48% | 3.33% | 22.54% | 57 | 0.392638 |
| both | 284 | 2.15% | 14.77% | 100.00% | 163 | 1.74233 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 353 | 2.56% | 16.30% | 84.65% | 106 | 2.16564 |
| anti | 64 | 0.46% | 2.96% | 15.35% | 57 | 0.392638 |
| both | 417 | 3.03% | 19.26% | 100.00% | 163 | 2.55828 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
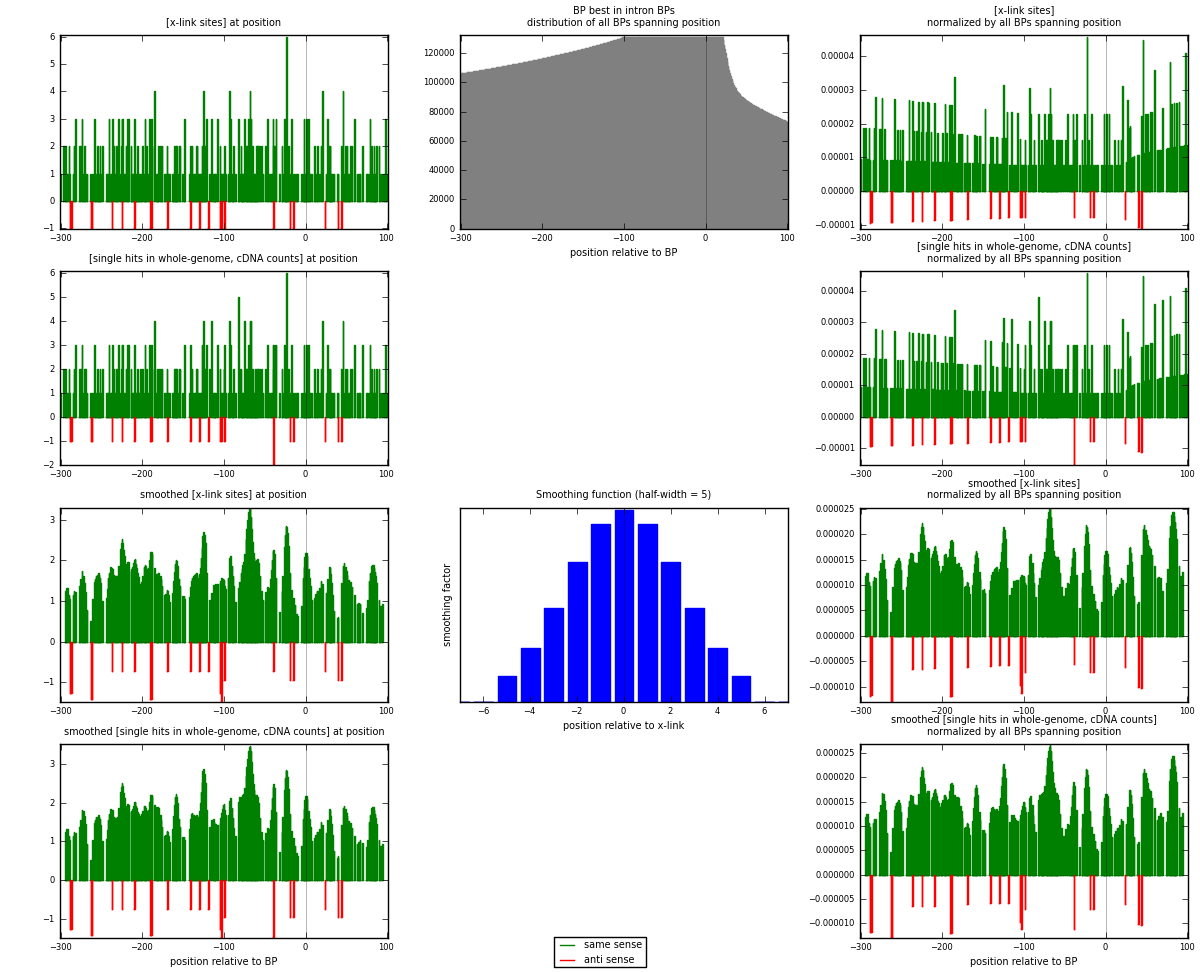 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 412 | 3.12% | 21.42% | 94.93% | 375 | 1.0404 |
| anti | 22 | 0.17% | 1.14% | 5.07% | 21 | 0.0555556 |
| both | 434 | 3.28% | 22.57% | 100.00% | 396 | 1.09596 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 430 | 3.12% | 19.86% | 94.92% | 375 | 1.08586 |
| anti | 23 | 0.17% | 1.06% | 5.08% | 21 | 0.0580808 |
| both | 453 | 3.29% | 20.92% | 100.00% | 396 | 1.14394 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.