RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
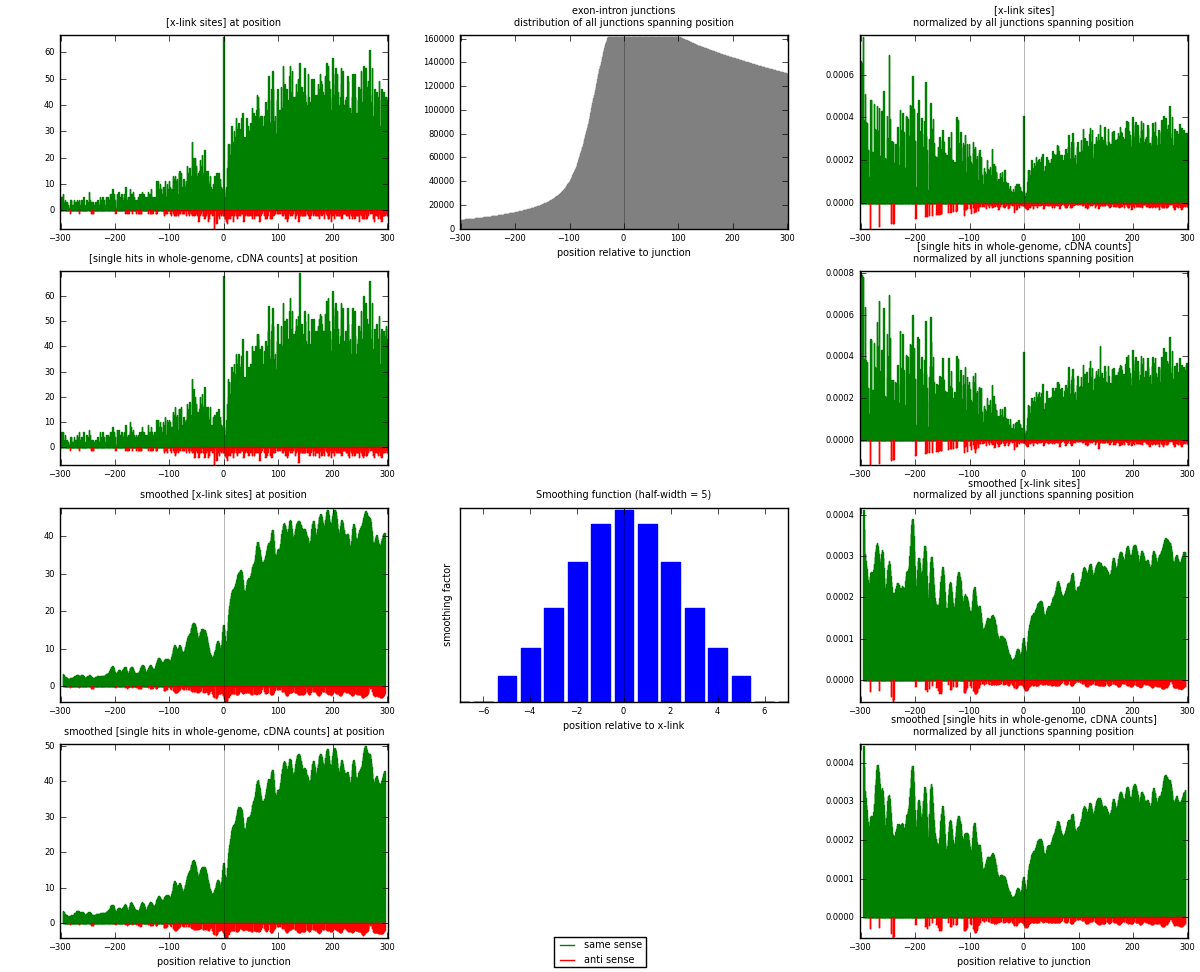 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 13408 | 4.17% | 27.20% | 96.30% | 9936 | 1.29408 |
| anti | 515 | 0.16% | 1.04% | 3.70% | 449 | 0.0497056 |
| both | 13923 | 4.33% | 28.24% | 100.00% | 10361 | 1.34379 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 14053 | 4.11% | 26.13% | 96.31% | 9936 | 1.35634 |
| anti | 538 | 0.16% | 1.00% | 3.69% | 449 | 0.0519255 |
| both | 14591 | 4.27% | 27.13% | 100.00% | 10361 | 1.40826 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
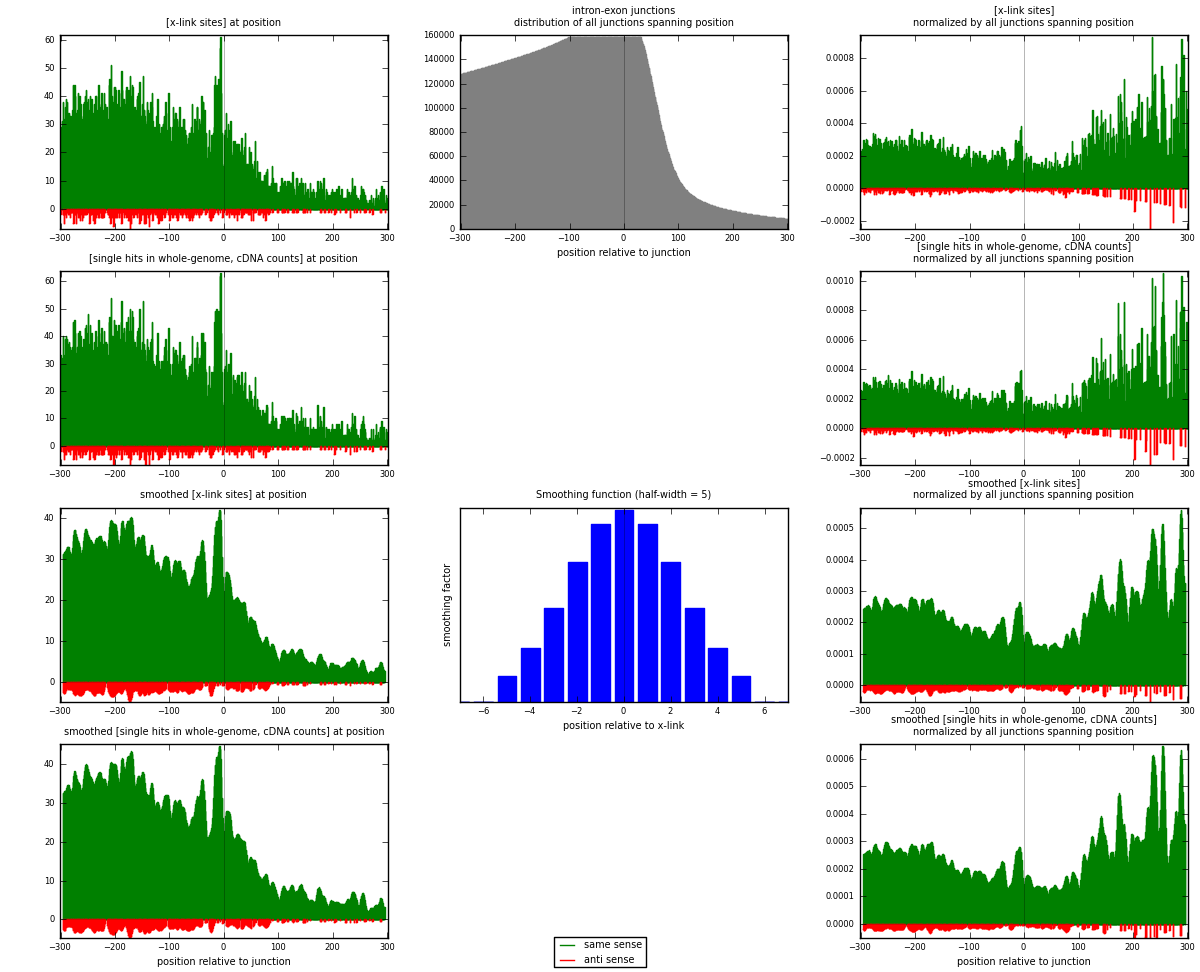 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 12103 | 3.76% | 24.55% | 94.93% | 9237 | 1.24146 |
| anti | 646 | 0.20% | 1.31% | 5.07% | 541 | 0.0662632 |
| both | 12749 | 3.96% | 25.86% | 100.00% | 9749 | 1.30772 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 12720 | 3.72% | 23.65% | 94.98% | 9237 | 1.30475 |
| anti | 672 | 0.20% | 1.25% | 5.02% | 541 | 0.0689301 |
| both | 13392 | 3.92% | 24.90% | 100.00% | 9749 | 1.37368 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
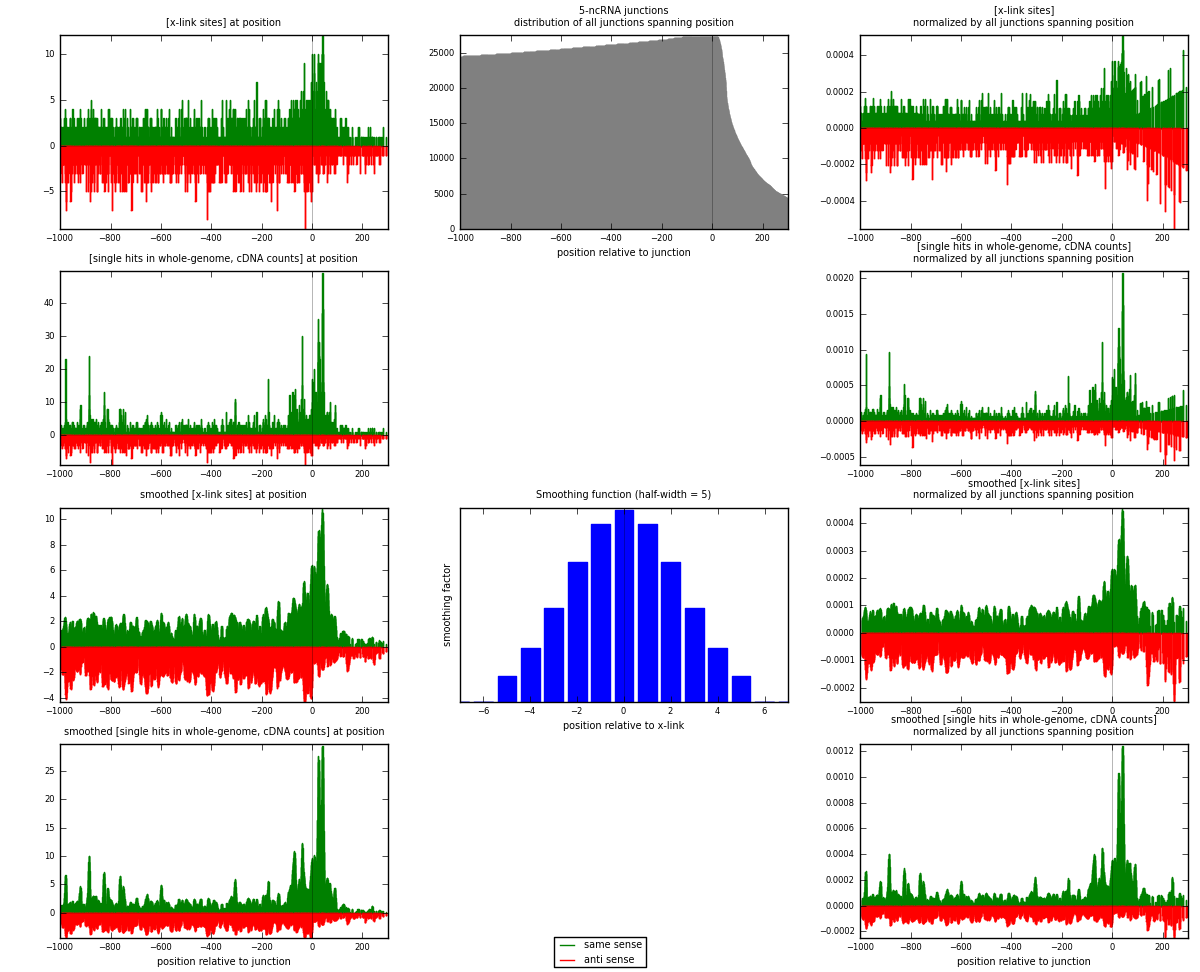 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1989 | 0.62% | 4.03% | 48.24% | 934 | 0.918707 |
| anti | 2134 | 0.66% | 4.33% | 51.76% | 1260 | 0.985681 |
| both | 4123 | 1.28% | 8.36% | 100.00% | 2165 | 1.90439 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2944 | 0.86% | 5.47% | 56.79% | 934 | 1.35982 |
| anti | 2240 | 0.66% | 4.17% | 43.21% | 1260 | 1.03464 |
| both | 5184 | 1.52% | 9.64% | 100.00% | 2165 | 2.39446 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
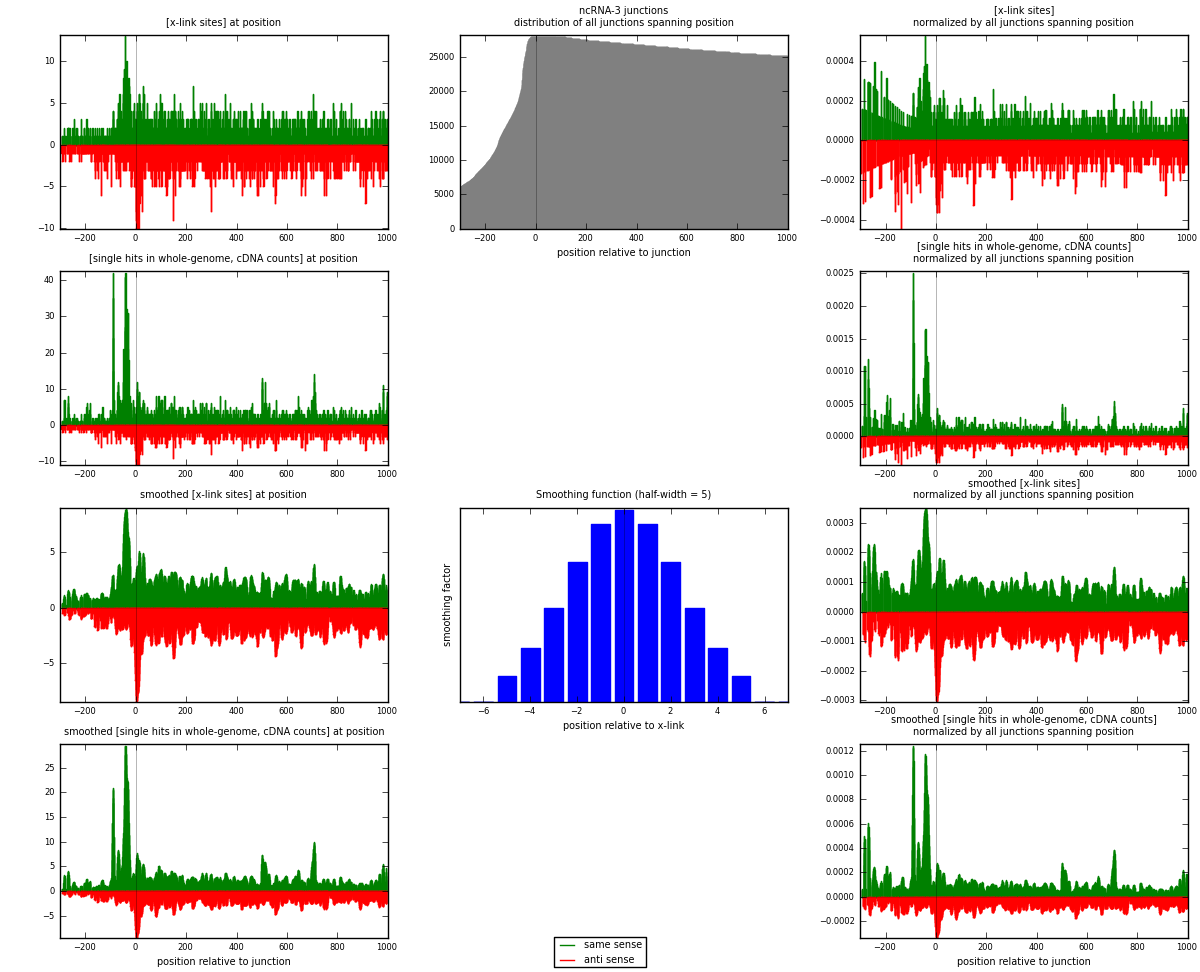 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2191 | 0.68% | 4.44% | 49.22% | 1004 | 0.952195 |
| anti | 2260 | 0.70% | 4.58% | 50.78% | 1315 | 0.982182 |
| both | 4451 | 1.38% | 9.03% | 100.00% | 2301 | 1.93438 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3109 | 0.91% | 5.78% | 56.47% | 1004 | 1.35115 |
| anti | 2397 | 0.70% | 4.46% | 43.53% | 1315 | 1.04172 |
| both | 5506 | 1.61% | 10.24% | 100.00% | 2301 | 2.39287 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
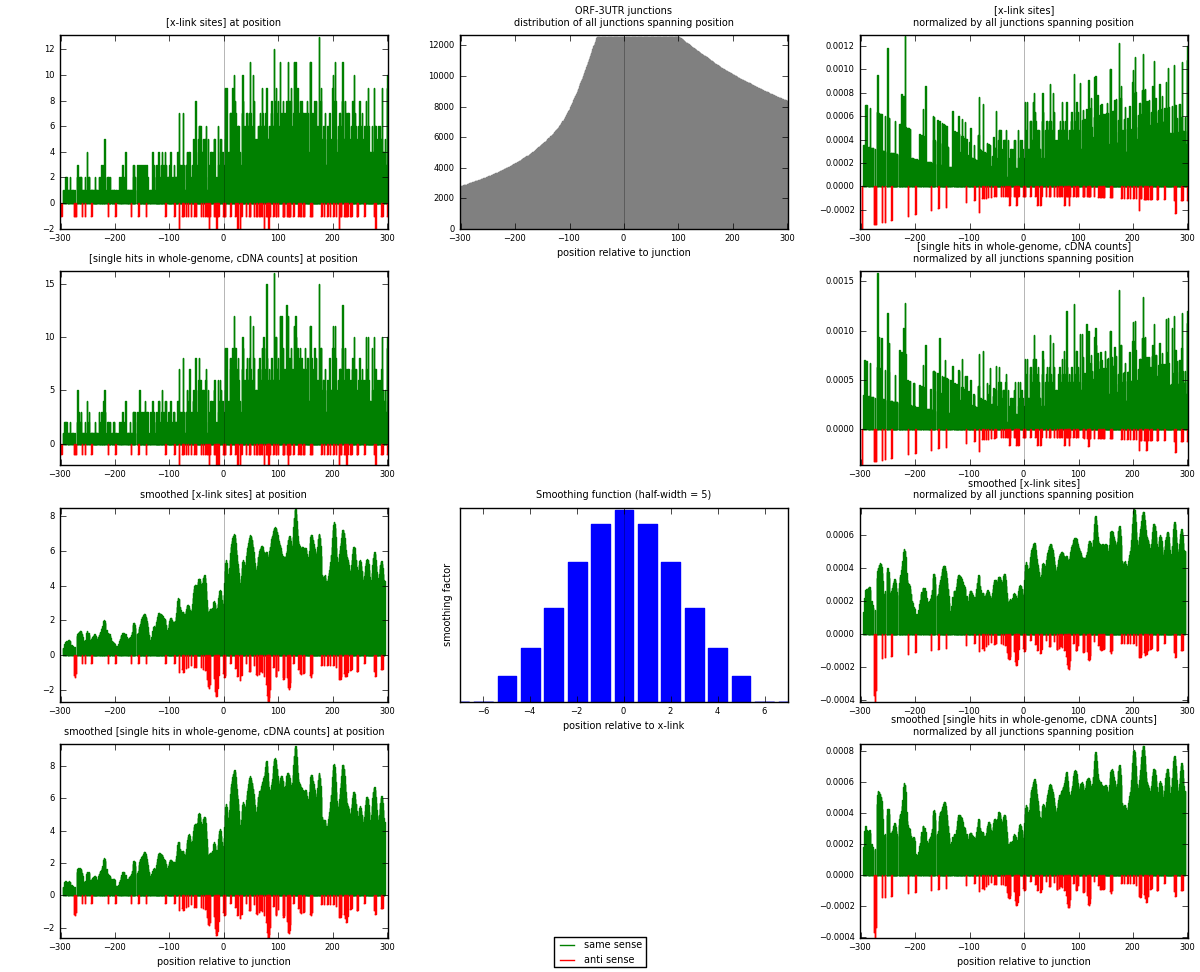 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2214 | 0.69% | 4.49% | 95.18% | 1244 | 1.66092 |
| anti | 112 | 0.03% | 0.23% | 4.82% | 97 | 0.084021 |
| both | 2326 | 0.72% | 4.72% | 100.00% | 1333 | 1.74494 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2407 | 0.70% | 4.48% | 95.44% | 1244 | 1.8057 |
| anti | 115 | 0.03% | 0.21% | 4.56% | 97 | 0.0862716 |
| both | 2522 | 0.74% | 4.69% | 100.00% | 1333 | 1.89197 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
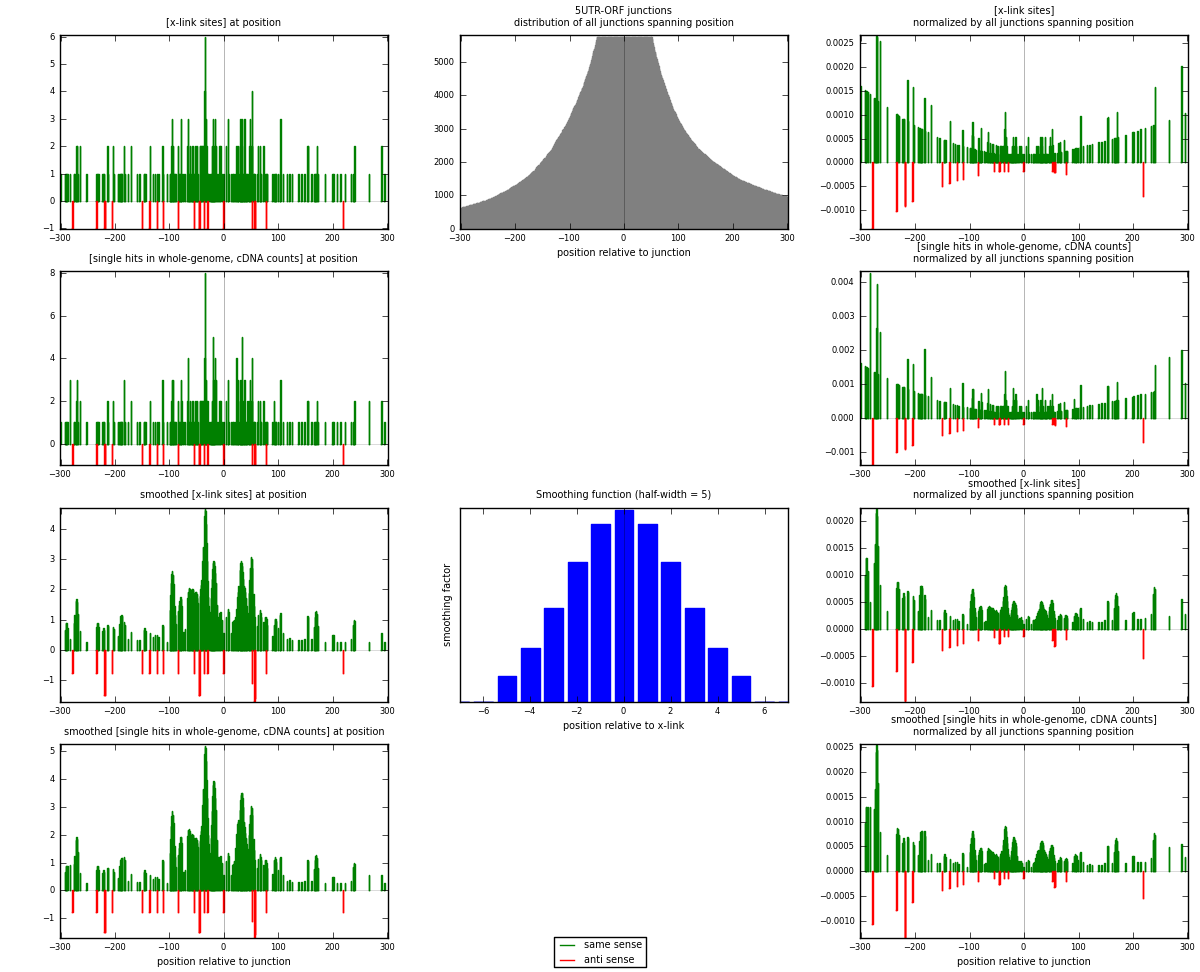 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 277 | 0.09% | 0.56% | 92.95% | 174 | 1.45789 |
| anti | 21 | 0.01% | 0.04% | 7.05% | 16 | 0.110526 |
| both | 298 | 0.09% | 0.60% | 100.00% | 190 | 1.56842 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 302 | 0.09% | 0.56% | 93.50% | 174 | 1.58947 |
| anti | 21 | 0.01% | 0.04% | 6.50% | 16 | 0.110526 |
| both | 323 | 0.09% | 0.60% | 100.00% | 190 | 1.7 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
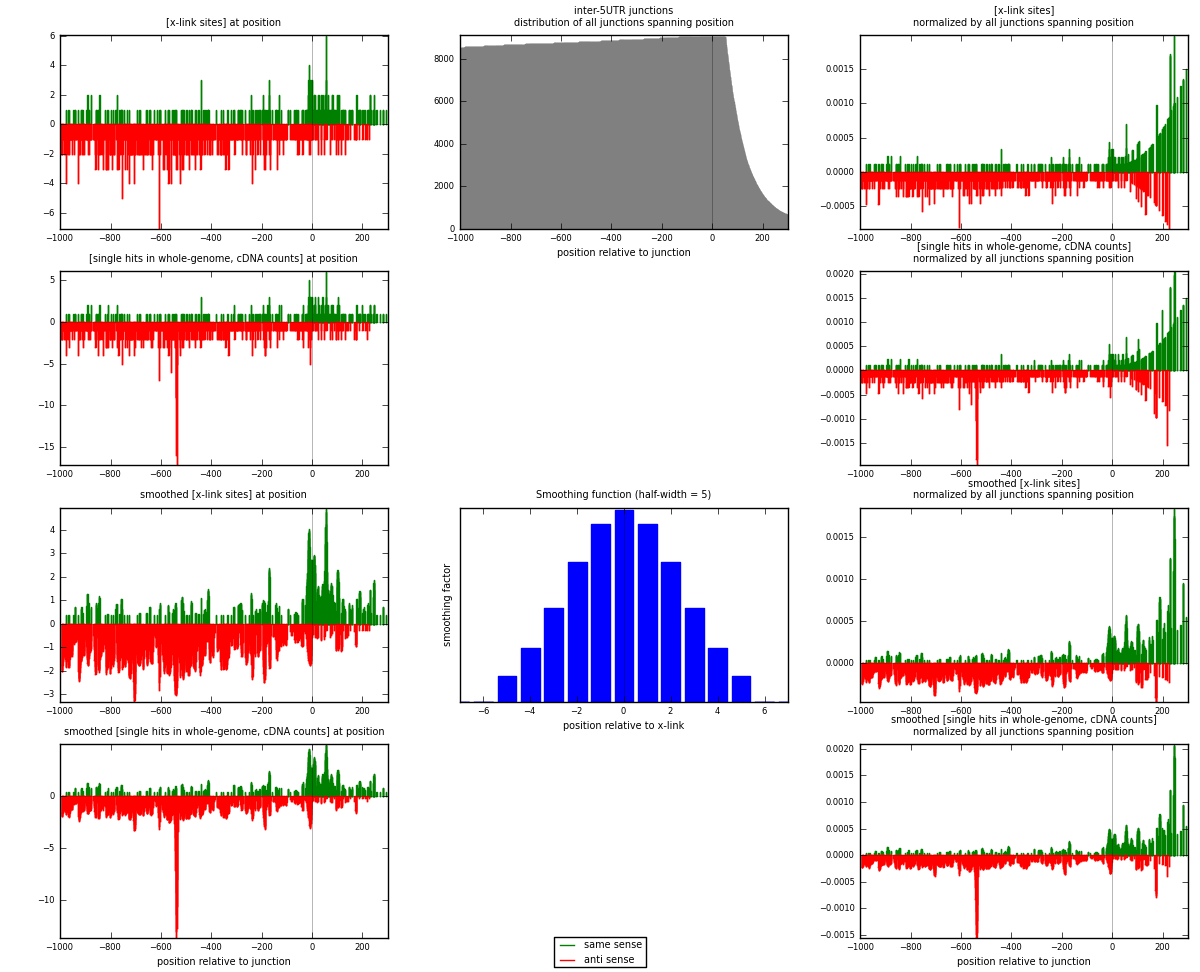 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 313 | 0.10% | 0.63% | 27.01% | 221 | 0.427012 |
| anti | 846 | 0.26% | 1.72% | 72.99% | 533 | 1.15416 |
| both | 1159 | 0.36% | 2.35% | 100.00% | 733 | 1.58117 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 330 | 0.10% | 0.61% | 25.90% | 221 | 0.450205 |
| anti | 944 | 0.28% | 1.76% | 74.10% | 533 | 1.28786 |
| both | 1274 | 0.37% | 2.37% | 100.00% | 733 | 1.73806 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
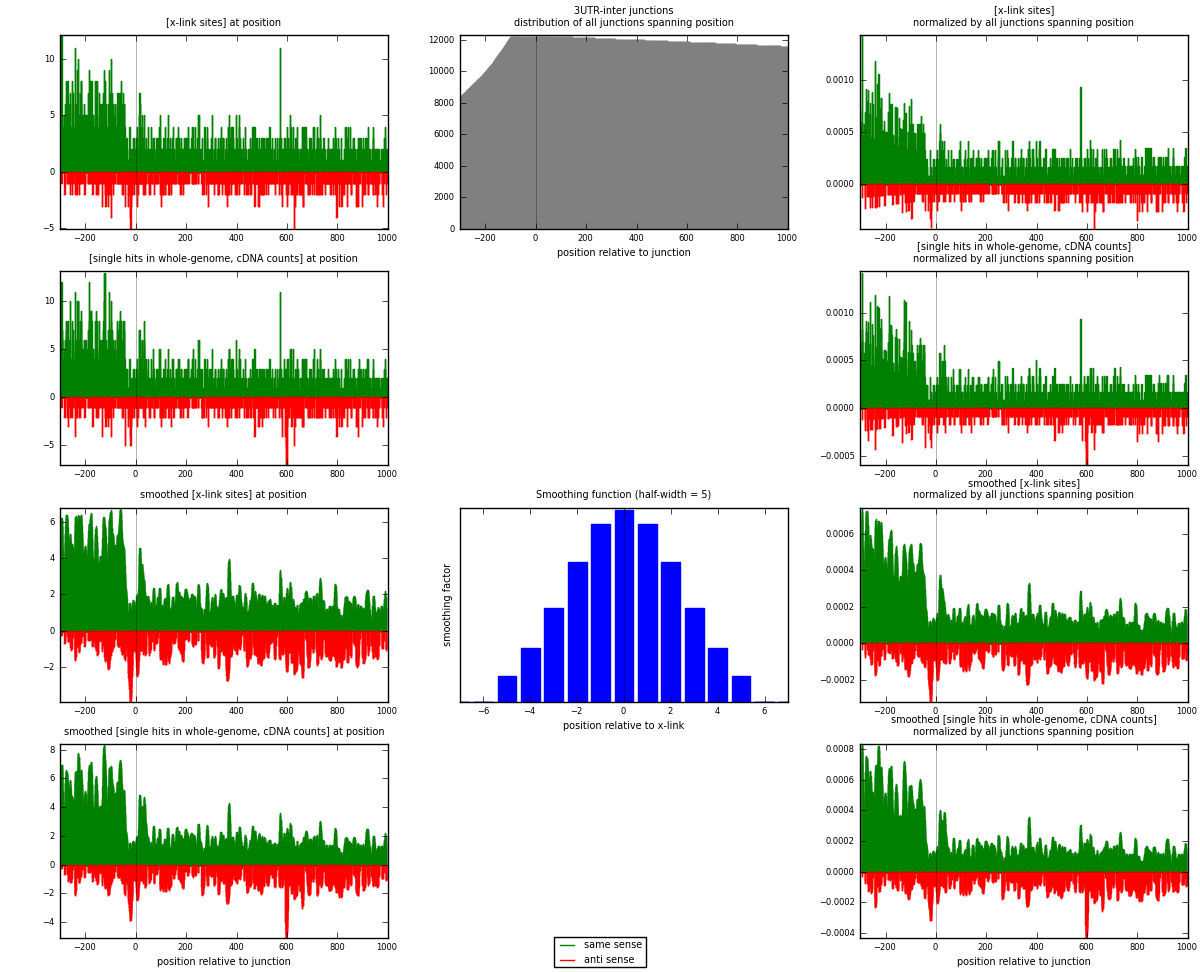 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2567 | 0.80% | 5.21% | 75.63% | 1560 | 1.27775 |
| anti | 827 | 0.26% | 1.68% | 24.37% | 507 | 0.411648 |
| both | 3394 | 1.05% | 6.88% | 100.00% | 2009 | 1.6894 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2741 | 0.80% | 5.10% | 75.61% | 1560 | 1.36436 |
| anti | 884 | 0.26% | 1.64% | 24.39% | 507 | 0.44002 |
| both | 3625 | 1.06% | 6.74% | 100.00% | 2009 | 1.80438 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
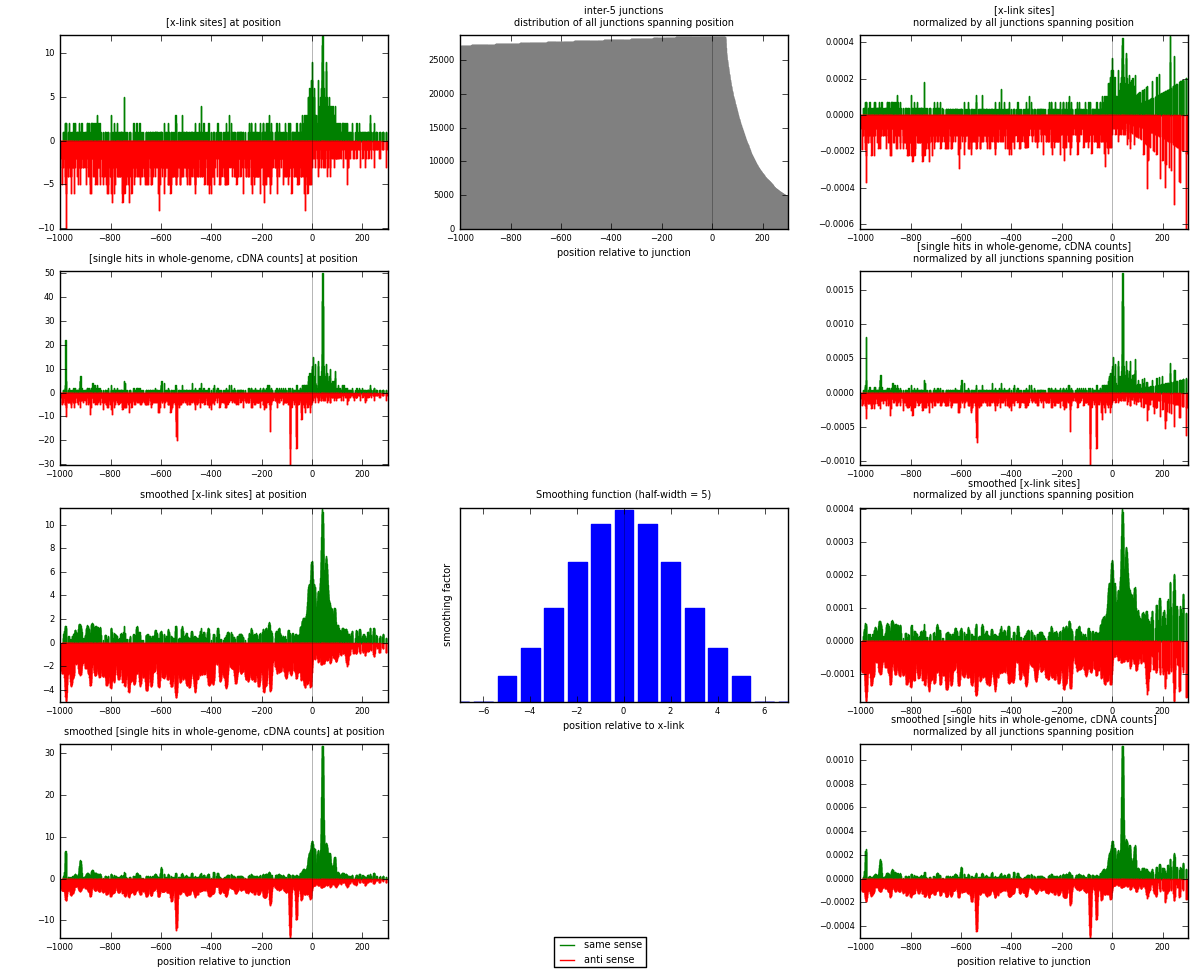 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 933 | 0.29% | 1.89% | 25.93% | 498 | 0.454678 |
| anti | 2665 | 0.83% | 5.41% | 74.07% | 1596 | 1.29873 |
| both | 3598 | 1.12% | 7.30% | 100.00% | 2052 | 1.75341 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1266 | 0.37% | 2.35% | 29.61% | 498 | 0.616959 |
| anti | 3010 | 0.88% | 5.60% | 70.39% | 1596 | 1.46686 |
| both | 4276 | 1.25% | 7.95% | 100.00% | 2052 | 2.08382 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
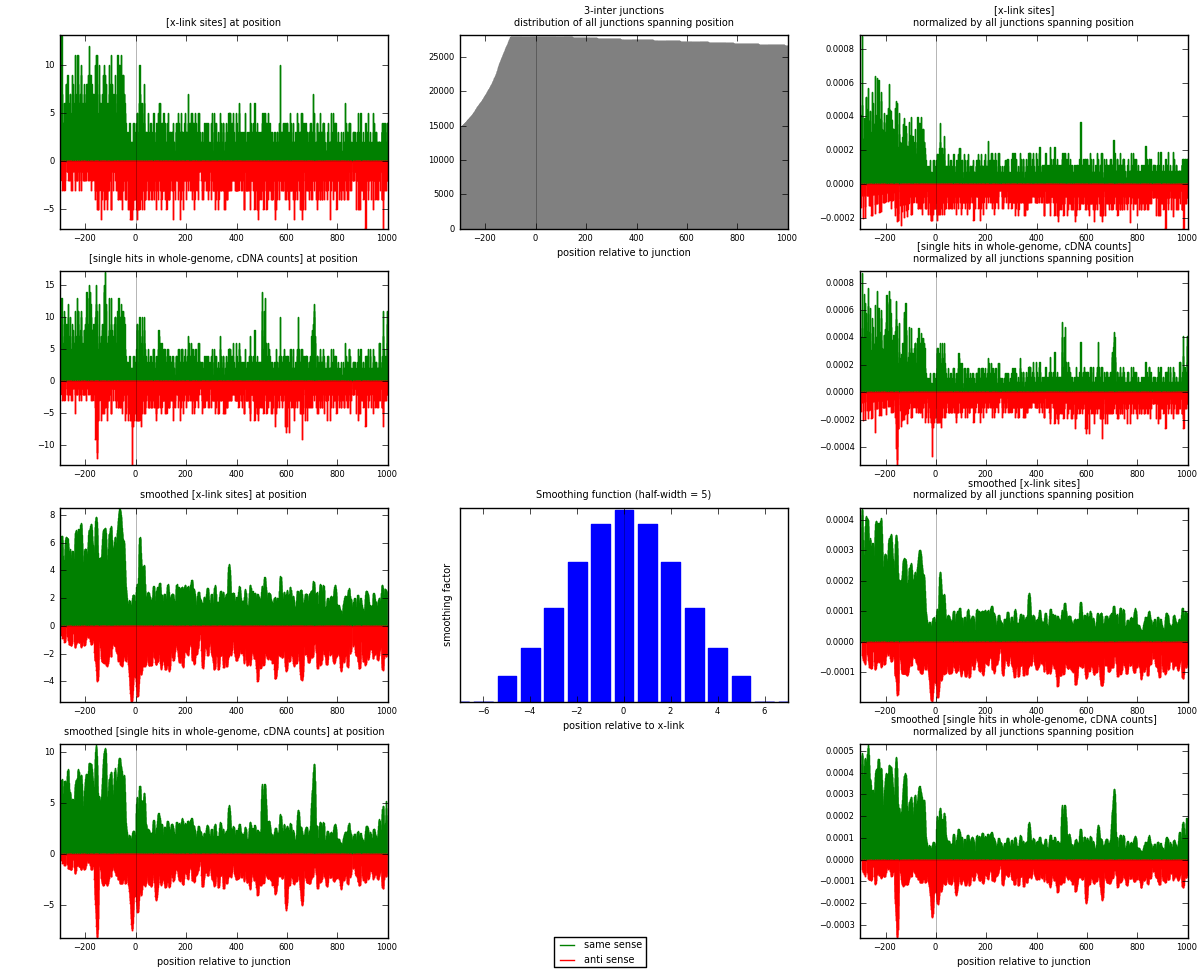 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3503 | 1.09% | 7.11% | 59.45% | 1967 | 1.08284 |
| anti | 2389 | 0.74% | 4.85% | 40.55% | 1345 | 0.738485 |
| both | 5892 | 1.83% | 11.95% | 100.00% | 3235 | 1.82133 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4108 | 1.20% | 7.64% | 61.18% | 1967 | 1.26986 |
| anti | 2607 | 0.76% | 4.85% | 38.82% | 1345 | 0.805873 |
| both | 6715 | 1.97% | 12.49% | 100.00% | 3235 | 2.07573 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
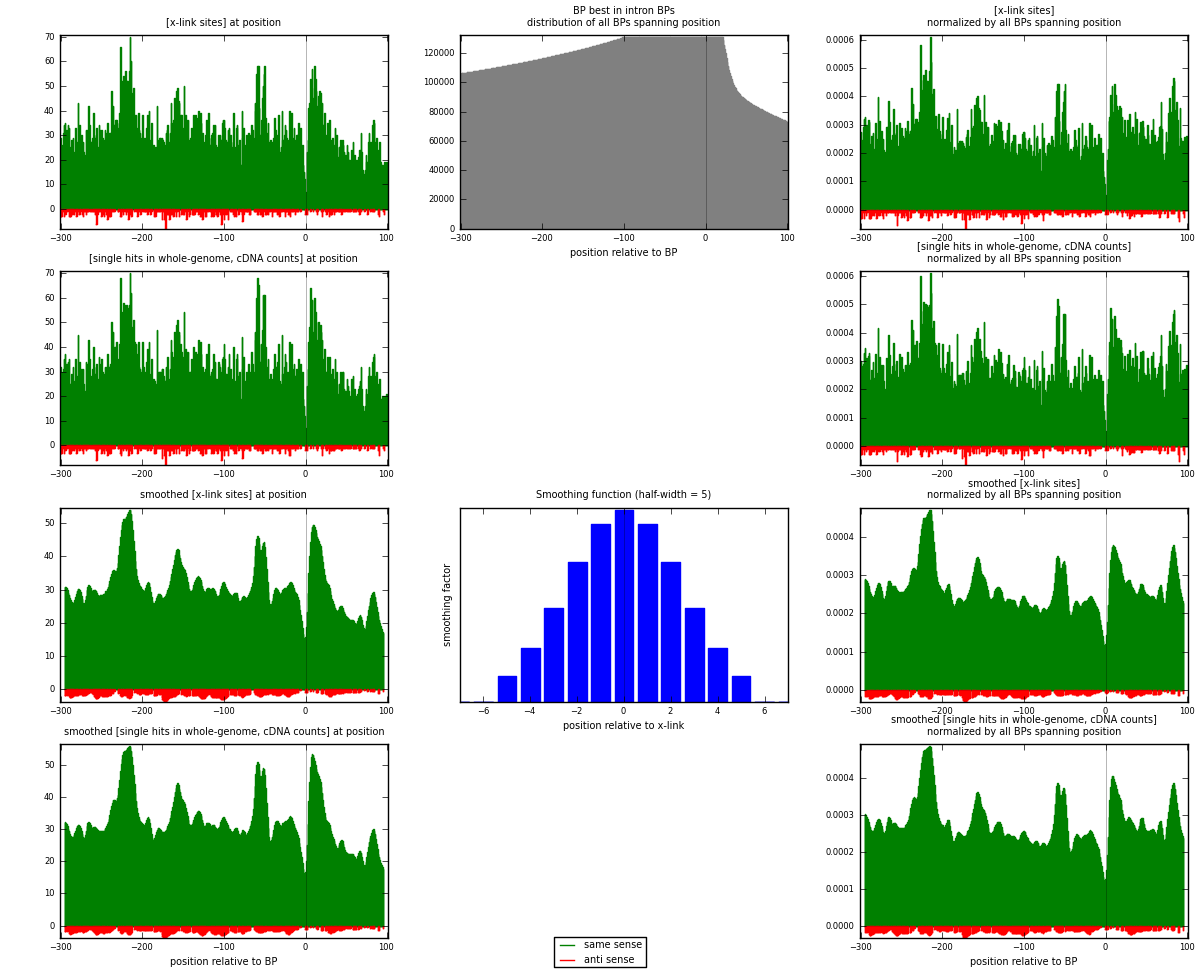 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 12339 | 3.83% | 25.03% | 96.27% | 9212 | 1.28411 |
| anti | 478 | 0.15% | 0.97% | 3.73% | 413 | 0.049745 |
| both | 12817 | 3.98% | 26.00% | 100.00% | 9609 | 1.33385 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 12960 | 3.79% | 24.10% | 96.39% | 9212 | 1.34874 |
| anti | 486 | 0.14% | 0.90% | 3.61% | 413 | 0.0505776 |
| both | 13446 | 3.93% | 25.00% | 100.00% | 9609 | 1.39931 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.