RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
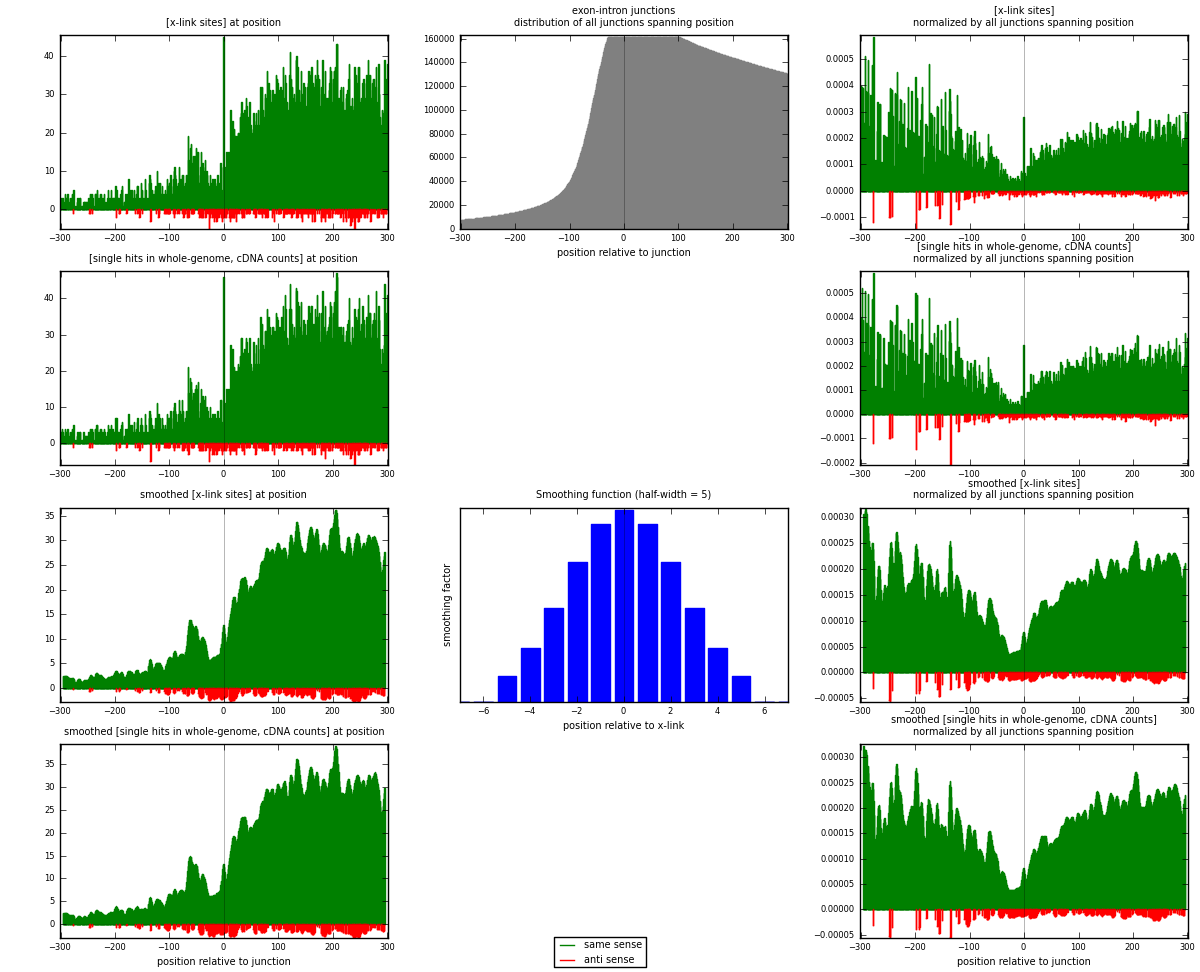 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 9466 | 4.05% | 26.47% | 96.31% | 7431 | 1.22284 |
| anti | 363 | 0.16% | 1.02% | 3.69% | 326 | 0.0468932 |
| both | 9829 | 4.21% | 27.49% | 100.00% | 7741 | 1.26973 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9941 | 4.00% | 25.39% | 96.29% | 7431 | 1.2842 |
| anti | 383 | 0.15% | 0.98% | 3.71% | 326 | 0.0494768 |
| both | 10324 | 4.16% | 26.36% | 100.00% | 7741 | 1.33368 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
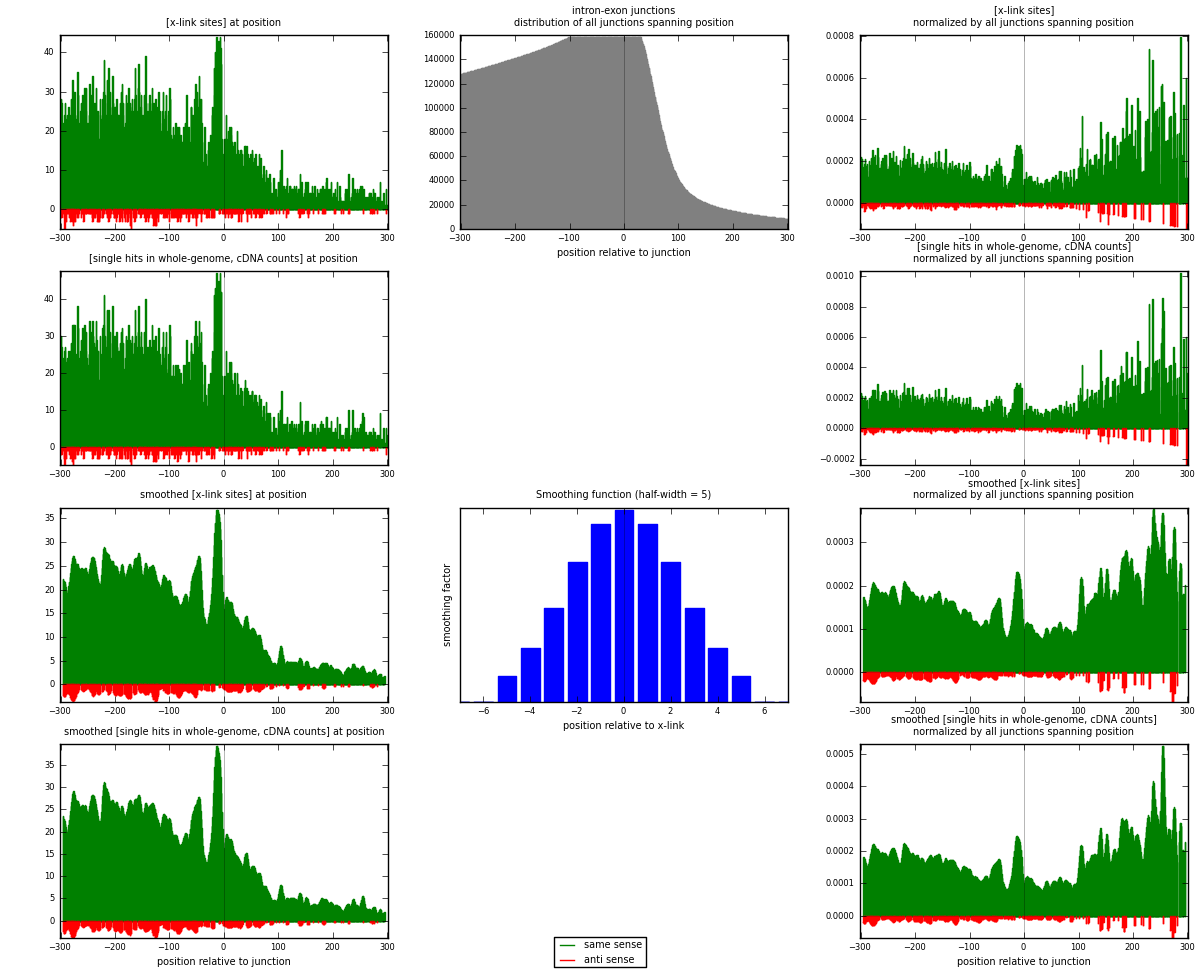 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 8785 | 3.76% | 24.57% | 95.14% | 6936 | 1.20129 |
| anti | 449 | 0.19% | 1.26% | 4.86% | 386 | 0.0613975 |
| both | 9234 | 3.95% | 25.82% | 100.00% | 7313 | 1.26268 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 9224 | 3.71% | 23.55% | 95.19% | 6936 | 1.26132 |
| anti | 466 | 0.19% | 1.19% | 4.81% | 386 | 0.0637221 |
| both | 9690 | 3.90% | 24.74% | 100.00% | 7313 | 1.32504 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
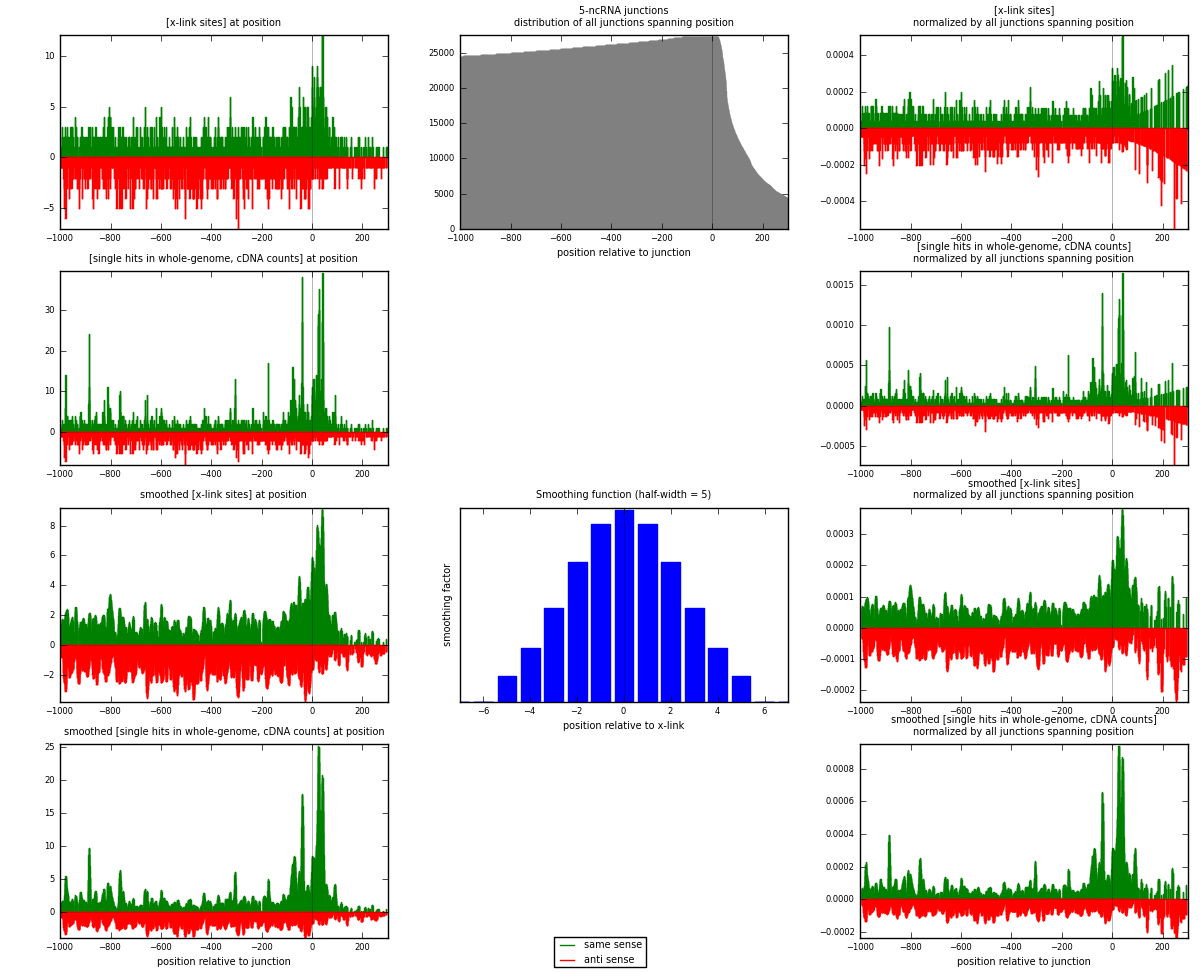 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1572 | 0.67% | 4.40% | 48.65% | 794 | 0.859486 |
| anti | 1659 | 0.71% | 4.64% | 51.35% | 1059 | 0.907053 |
| both | 3231 | 1.38% | 9.04% | 100.00% | 1829 | 1.76654 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2325 | 0.94% | 5.94% | 57.27% | 794 | 1.27119 |
| anti | 1735 | 0.70% | 4.43% | 42.73% | 1059 | 0.948606 |
| both | 4060 | 1.63% | 10.37% | 100.00% | 1829 | 2.21979 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
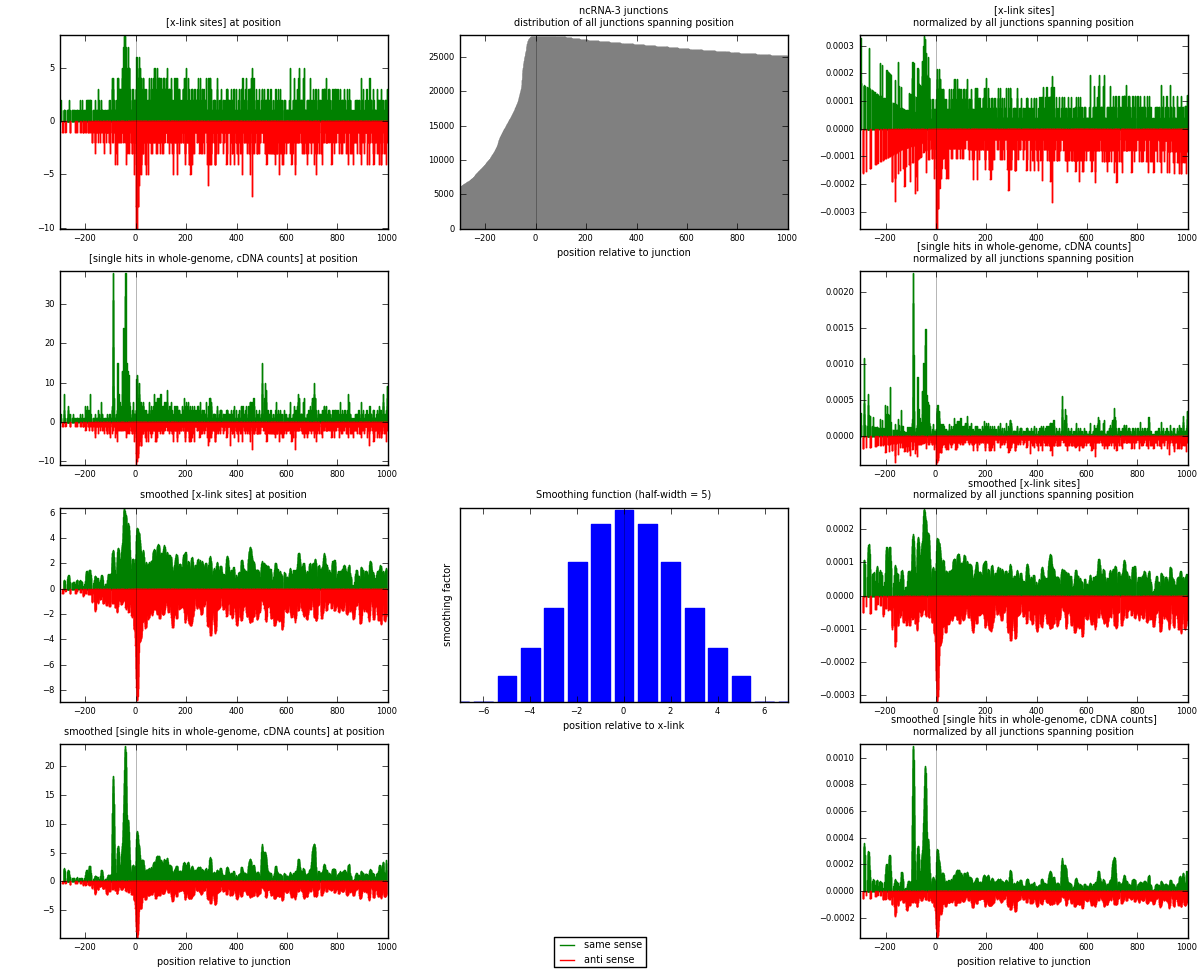 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1789 | 0.77% | 5.00% | 52.40% | 849 | 0.953117 |
| anti | 1625 | 0.70% | 4.54% | 47.60% | 1036 | 0.865743 |
| both | 3414 | 1.46% | 9.55% | 100.00% | 1877 | 1.81886 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2499 | 1.01% | 6.38% | 59.33% | 849 | 1.33138 |
| anti | 1713 | 0.69% | 4.37% | 40.67% | 1036 | 0.912627 |
| both | 4212 | 1.70% | 10.76% | 100.00% | 1877 | 2.24401 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
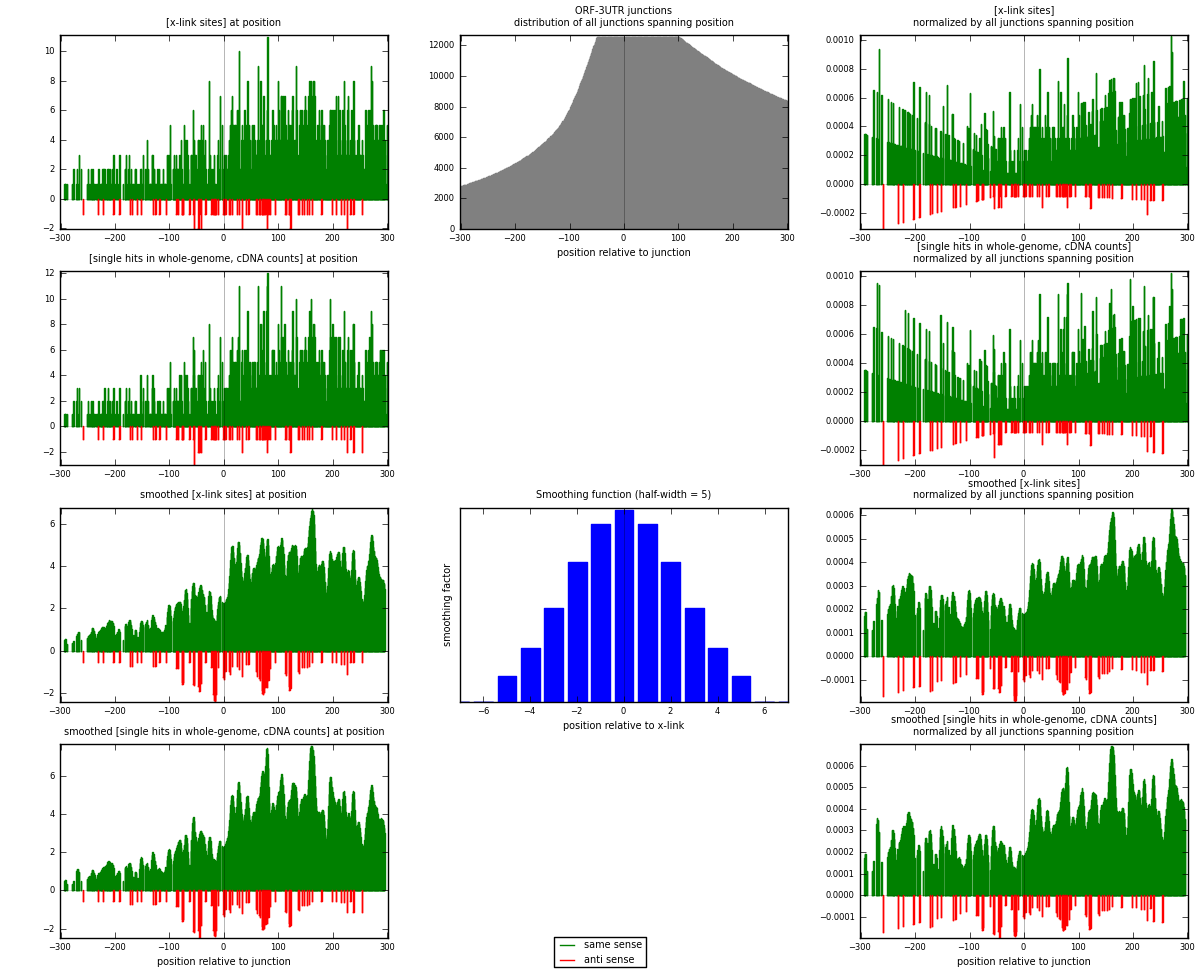
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1538 | 0.66% | 4.30% | 94.82% | 955 | 1.51081 |
| anti | 84 | 0.04% | 0.23% | 5.18% | 70 | 0.0825147 |
| both | 1622 | 0.69% | 4.54% | 100.00% | 1018 | 1.59332 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1653 | 0.67% | 4.22% | 94.95% | 955 | 1.62377 |
| anti | 88 | 0.04% | 0.22% | 5.05% | 70 | 0.086444 |
| both | 1741 | 0.70% | 4.45% | 100.00% | 1018 | 1.71022 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
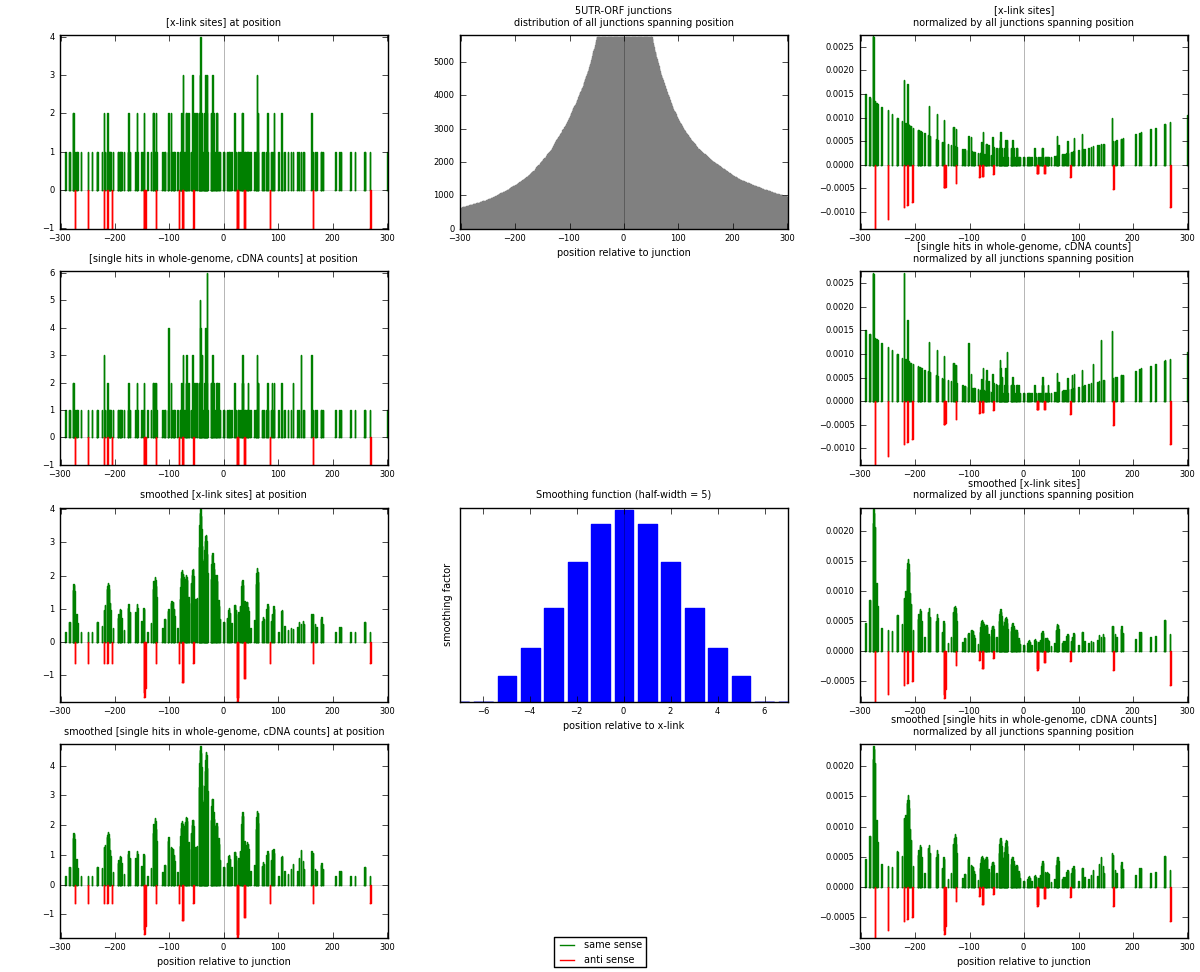
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 231 | 0.10% | 0.65% | 91.67% | 160 | 1.32759 |
| anti | 21 | 0.01% | 0.06% | 8.33% | 14 | 0.12069 |
| both | 252 | 0.11% | 0.70% | 100.00% | 174 | 1.44828 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 255 | 0.10% | 0.65% | 92.39% | 160 | 1.46552 |
| anti | 21 | 0.01% | 0.05% | 7.61% | 14 | 0.12069 |
| both | 276 | 0.11% | 0.70% | 100.00% | 174 | 1.58621 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
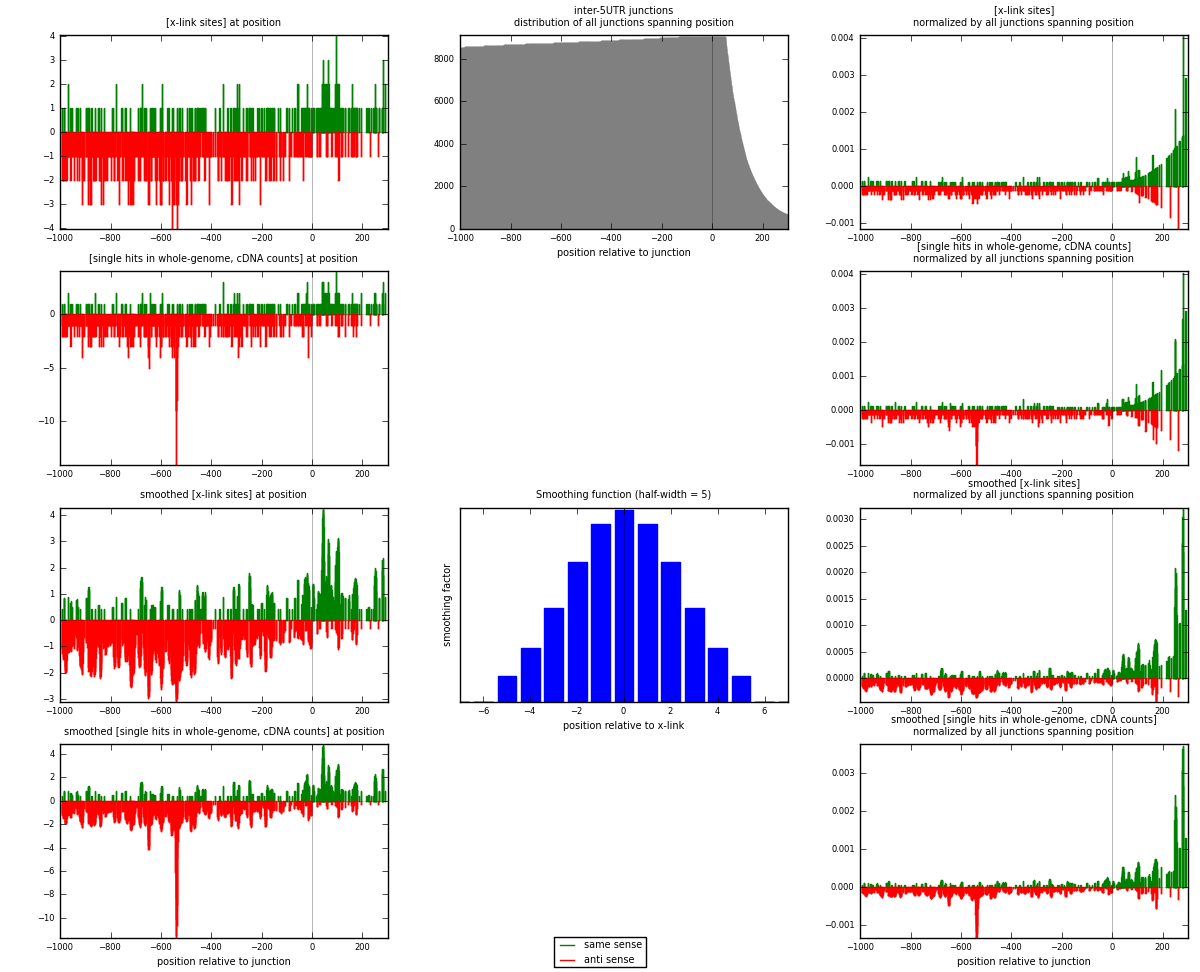
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 259 | 0.11% | 0.72% | 28.31% | 175 | 0.445017 |
| anti | 656 | 0.28% | 1.83% | 71.69% | 420 | 1.12715 |
| both | 915 | 0.39% | 2.56% | 100.00% | 582 | 1.57216 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 270 | 0.11% | 0.69% | 26.68% | 175 | 0.463918 |
| anti | 742 | 0.30% | 1.89% | 73.32% | 420 | 1.27491 |
| both | 1012 | 0.41% | 2.58% | 100.00% | 582 | 1.73883 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
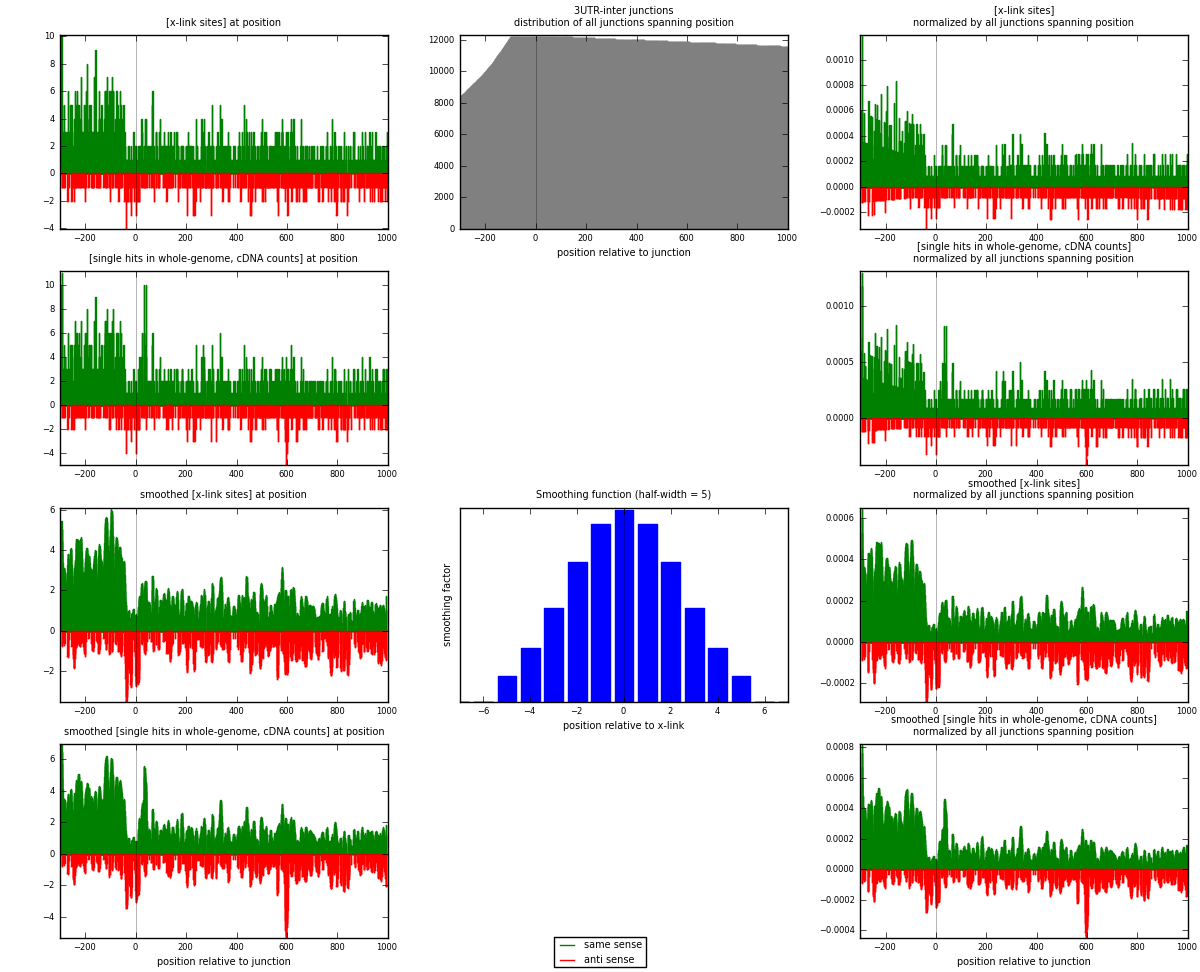
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1800 | 0.77% | 5.03% | 76.69% | 1160 | 1.19601 |
| anti | 547 | 0.23% | 1.53% | 23.31% | 373 | 0.363455 |
| both | 2347 | 1.00% | 6.56% | 100.00% | 1505 | 1.55947 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1944 | 0.78% | 4.96% | 76.84% | 1160 | 1.29169 |
| anti | 586 | 0.24% | 1.50% | 23.16% | 373 | 0.389369 |
| both | 2530 | 1.02% | 6.46% | 100.00% | 1505 | 1.68106 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
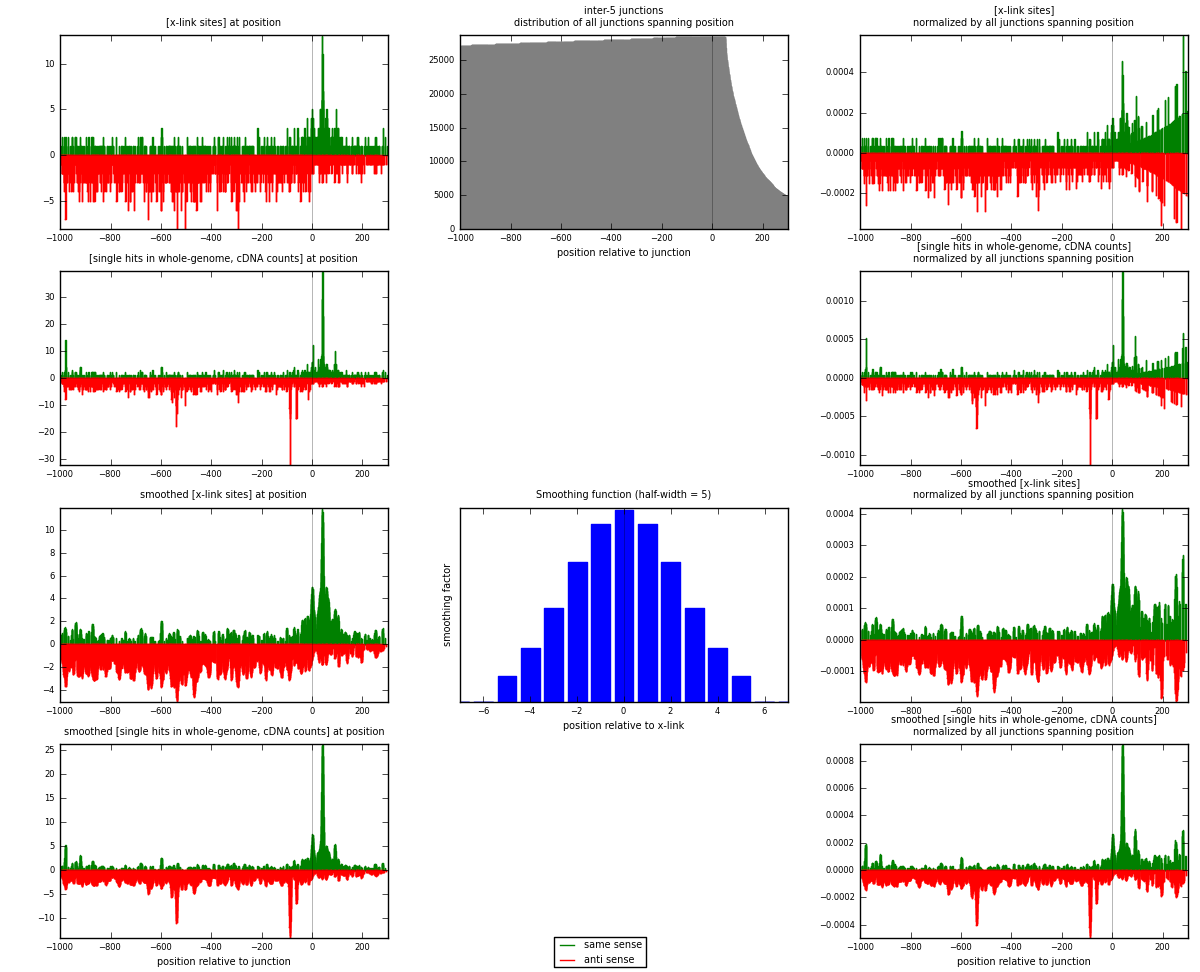
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 743 | 0.32% | 2.08% | 26.18% | 428 | 0.436289 |
| anti | 2095 | 0.90% | 5.86% | 73.82% | 1309 | 1.23018 |
| both | 2838 | 1.21% | 7.94% | 100.00% | 1703 | 1.66647 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 926 | 0.37% | 2.36% | 28.15% | 428 | 0.543746 |
| anti | 2363 | 0.95% | 6.03% | 71.85% | 1309 | 1.38755 |
| both | 3289 | 1.32% | 8.40% | 100.00% | 1703 | 1.9313 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
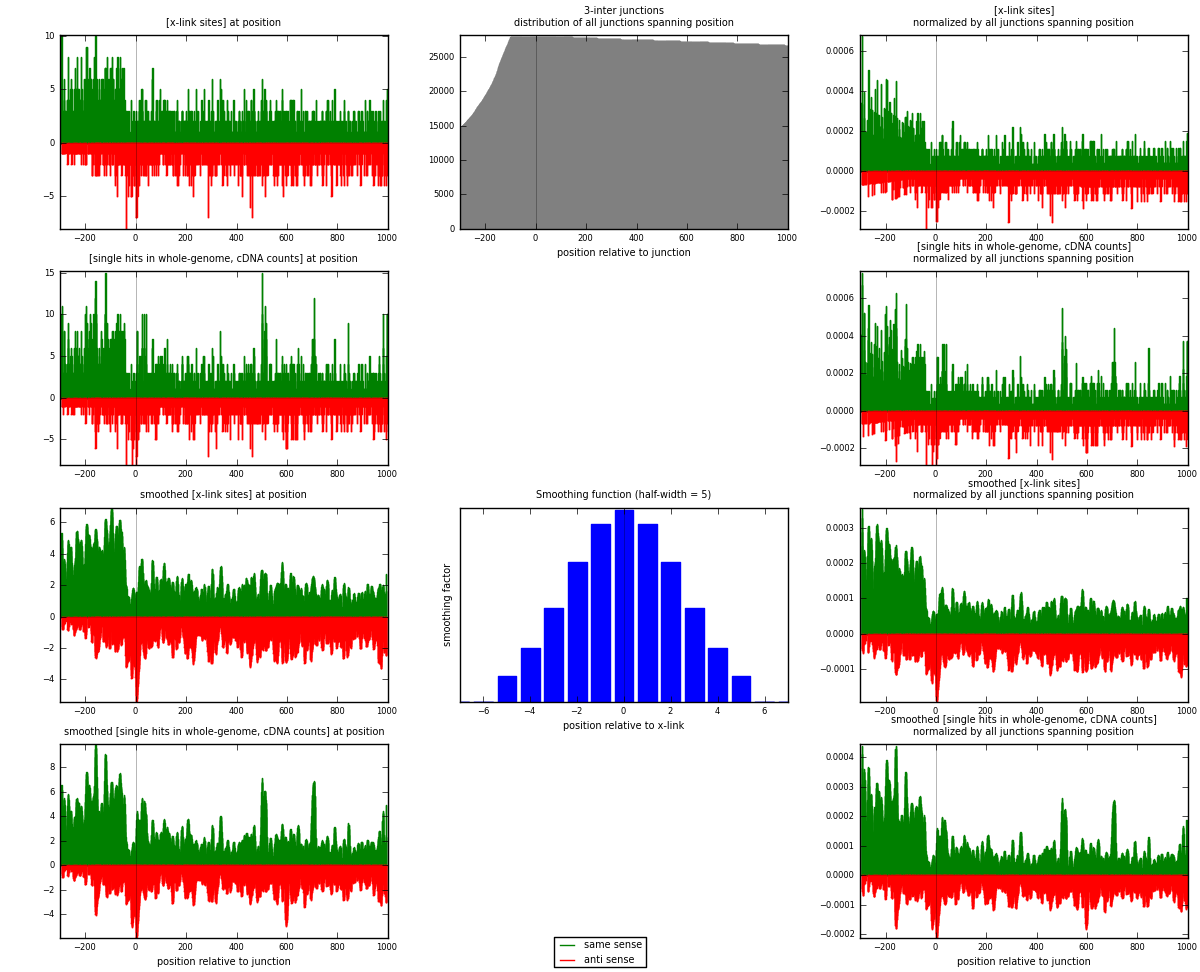
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 2560 | 1.10% | 7.16% | 60.33% | 1495 | 1.02277 |
| anti | 1683 | 0.72% | 4.71% | 39.67% | 1045 | 0.672393 |
| both | 4243 | 1.82% | 11.87% | 100.00% | 2503 | 1.69517 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3096 | 1.25% | 7.91% | 63.08% | 1495 | 1.23692 |
| anti | 1812 | 0.73% | 4.63% | 36.92% | 1045 | 0.723931 |
| both | 4908 | 1.98% | 12.53% | 100.00% | 2503 | 1.96085 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
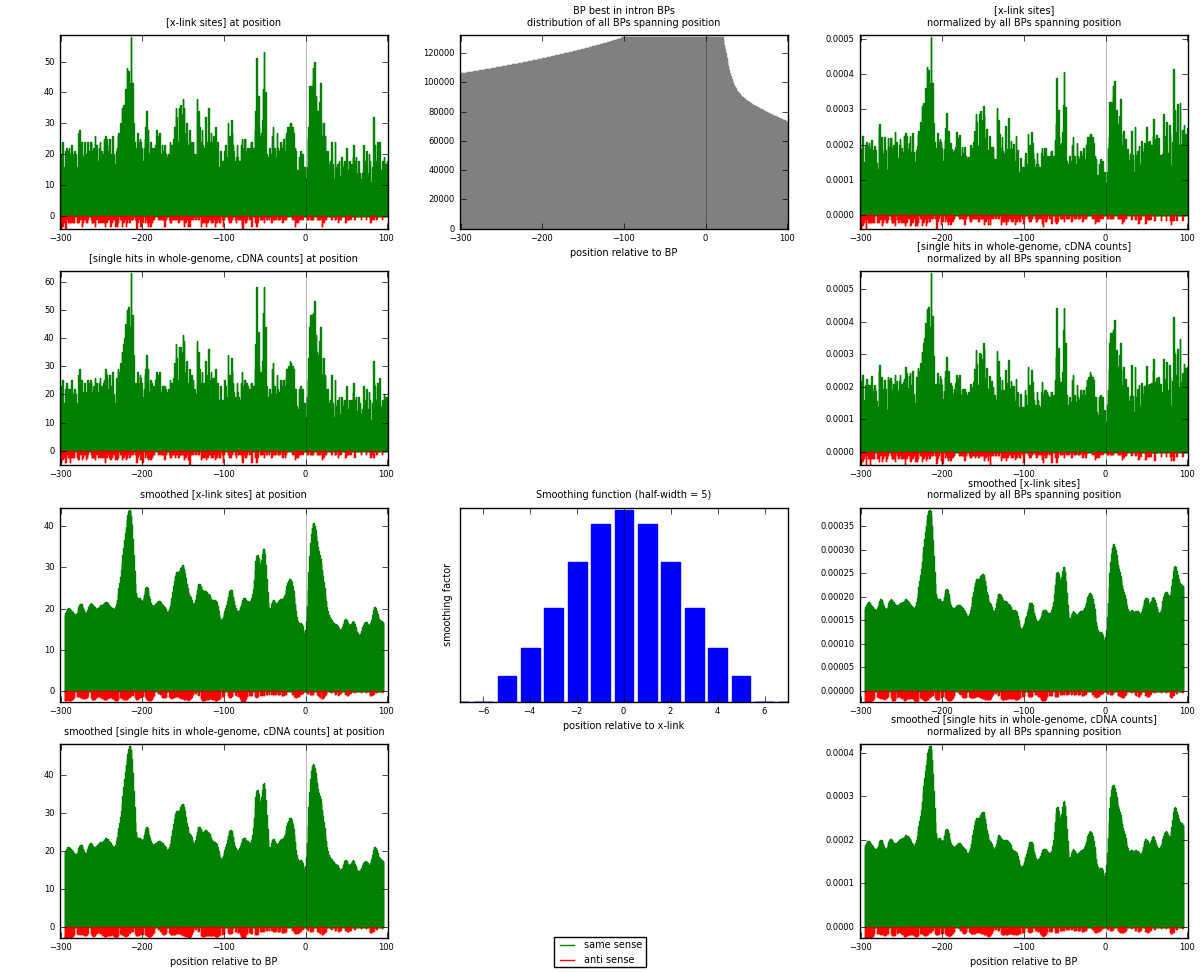
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 9026 | 3.86% | 25.24% | 96.21% | 7064 | 1.22469 |
| anti | 356 | 0.15% | 1.00% | 3.79% | 319 | 0.0483039 |
| both | 9382 | 4.02% | 26.24% | 100.00% | 7370 | 1.273 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 9486 | 3.82% | 24.22% | 96.22% | 7064 | 1.28711 |
| anti | 373 | 0.15% | 0.95% | 3.78% | 319 | 0.0506106 |
| both | 9859 | 3.97% | 25.18% | 100.00% | 7370 | 1.33772 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.