RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
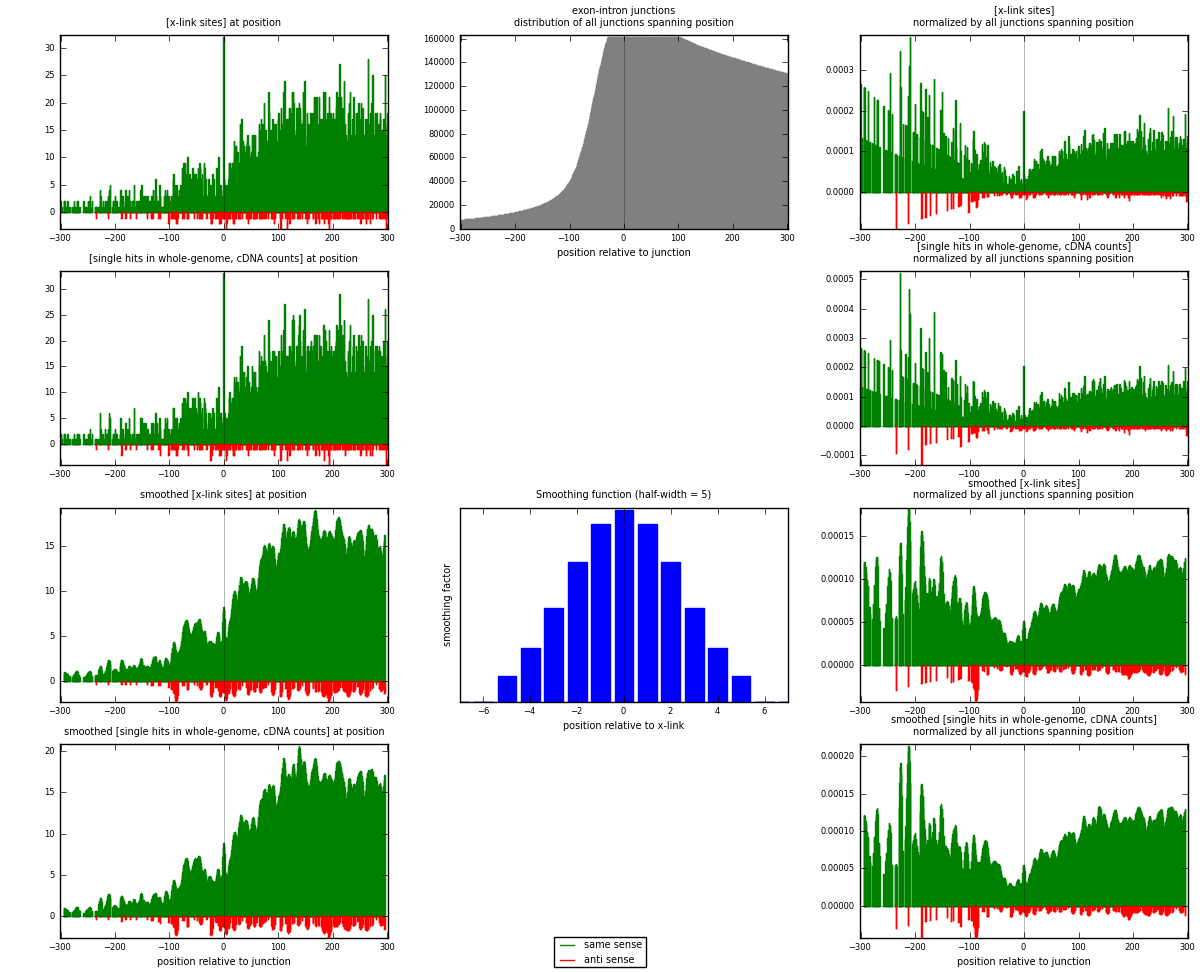 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 5041 | 4.02% | 26.14% | 96.00% | 4234 | 1.14308 |
| anti | 210 | 0.17% | 1.09% | 4.00% | 177 | 0.047619 |
| both | 5251 | 4.19% | 27.23% | 100.00% | 4410 | 1.1907 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 5284 | 4.00% | 25.26% | 96.00% | 4234 | 1.19819 |
| anti | 220 | 0.17% | 1.05% | 4.00% | 177 | 0.0498866 |
| both | 5504 | 4.16% | 26.31% | 100.00% | 4410 | 1.24807 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
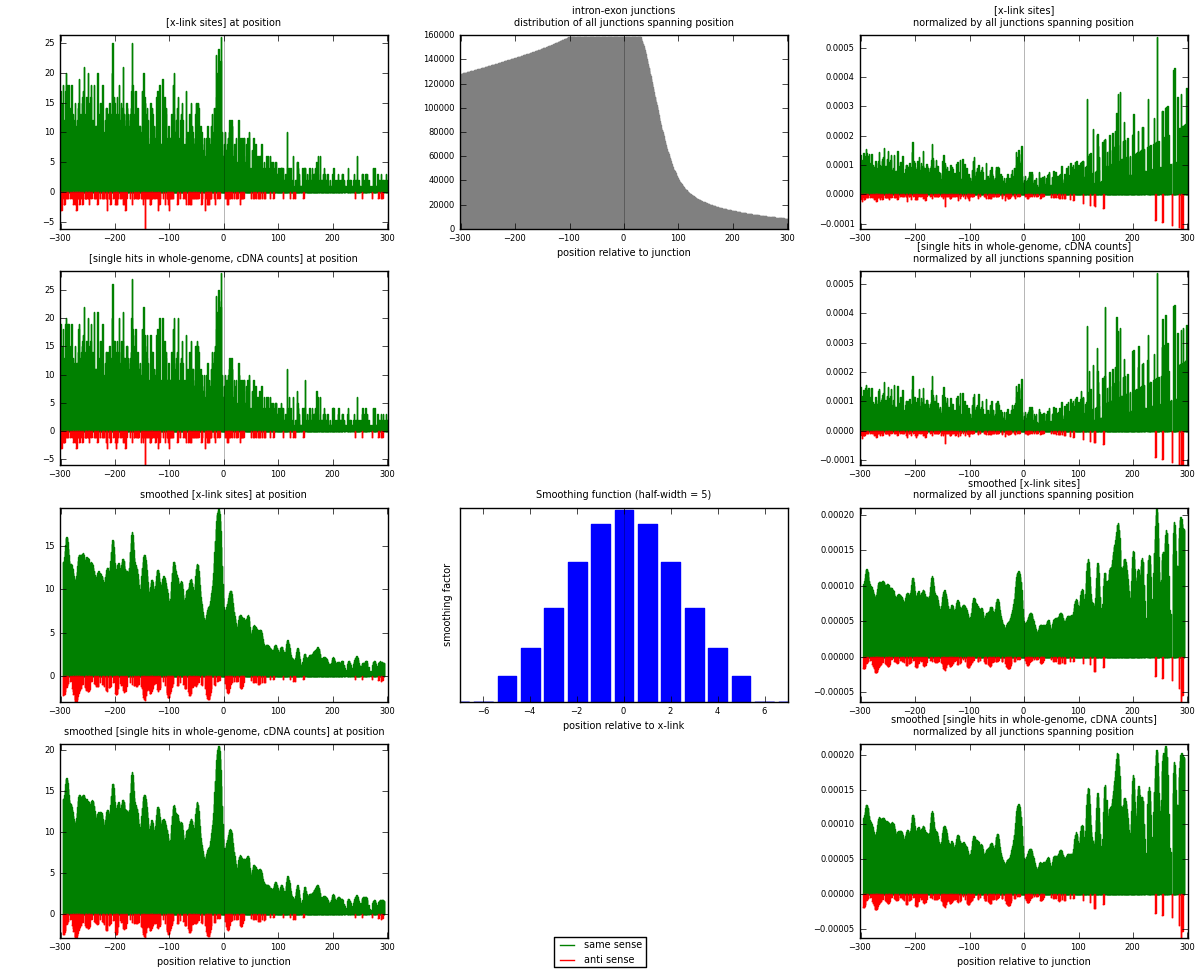 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4500 | 3.59% | 23.34% | 95.12% | 3856 | 1.11001 |
| anti | 231 | 0.18% | 1.20% | 4.88% | 205 | 0.0569808 |
| both | 4731 | 3.78% | 24.54% | 100.00% | 4054 | 1.167 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4698 | 3.55% | 22.46% | 95.22% | 3856 | 1.15886 |
| anti | 236 | 0.18% | 1.13% | 4.78% | 205 | 0.0582141 |
| both | 4934 | 3.73% | 23.59% | 100.00% | 4054 | 1.21707 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
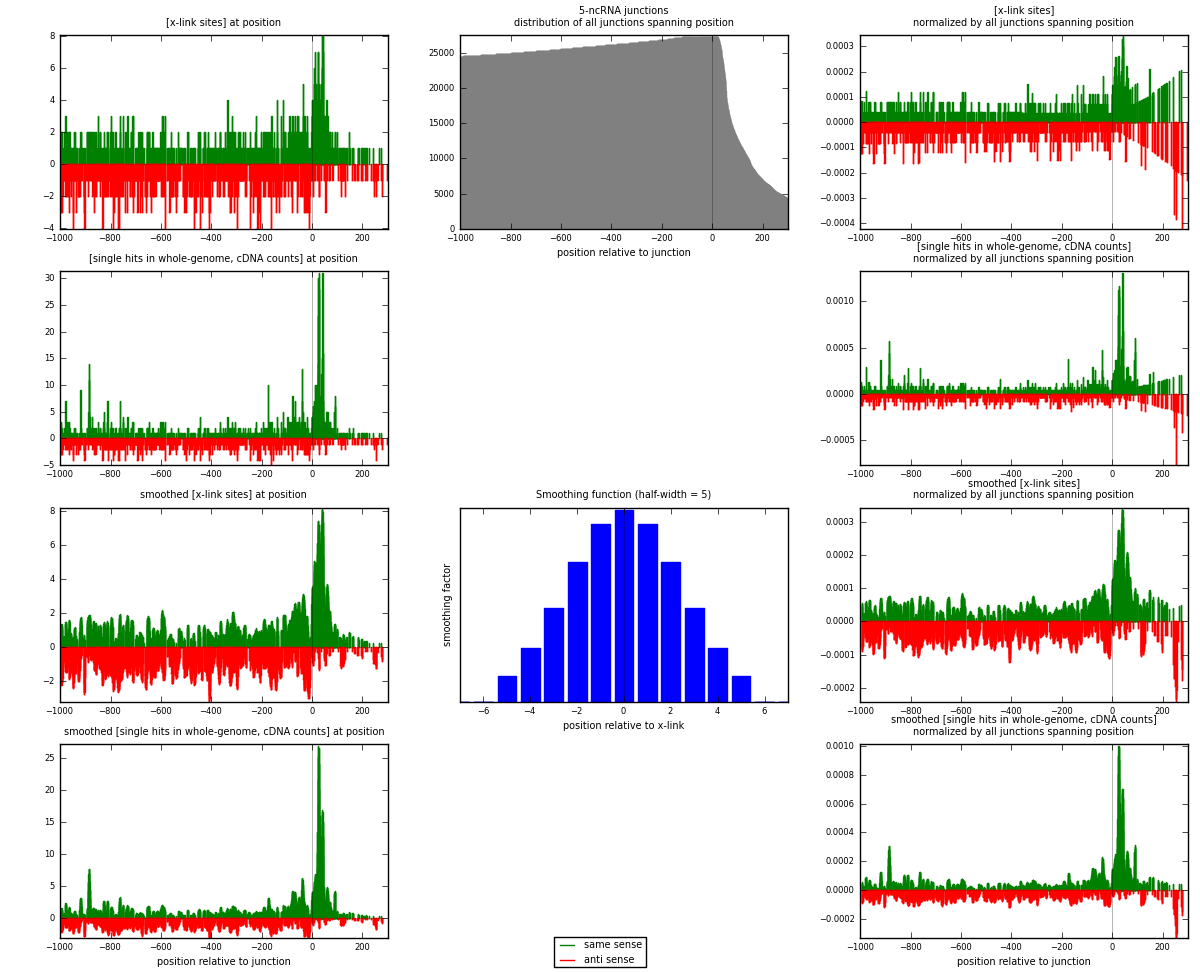 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 965 | 0.77% | 5.00% | 51.80% | 519 | 0.813659 |
| anti | 898 | 0.72% | 4.66% | 48.20% | 673 | 0.757167 |
| both | 1863 | 1.49% | 9.66% | 100.00% | 1186 | 1.57083 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1352 | 1.02% | 6.46% | 58.78% | 519 | 1.13997 |
| anti | 948 | 0.72% | 4.53% | 41.22% | 673 | 0.799325 |
| both | 2300 | 1.74% | 10.99% | 100.00% | 1186 | 1.93929 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1027 | 0.82% | 5.33% | 53.60% | 513 | 0.880789 |
| anti | 889 | 0.71% | 4.61% | 46.40% | 657 | 0.762436 |
| both | 1916 | 1.53% | 9.94% | 100.00% | 1166 | 1.64322 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1374 | 1.04% | 6.57% | 59.79% | 513 | 1.17839 |
| anti | 924 | 0.70% | 4.42% | 40.21% | 657 | 0.792453 |
| both | 2298 | 1.74% | 10.98% | 100.00% | 1166 | 1.97084 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
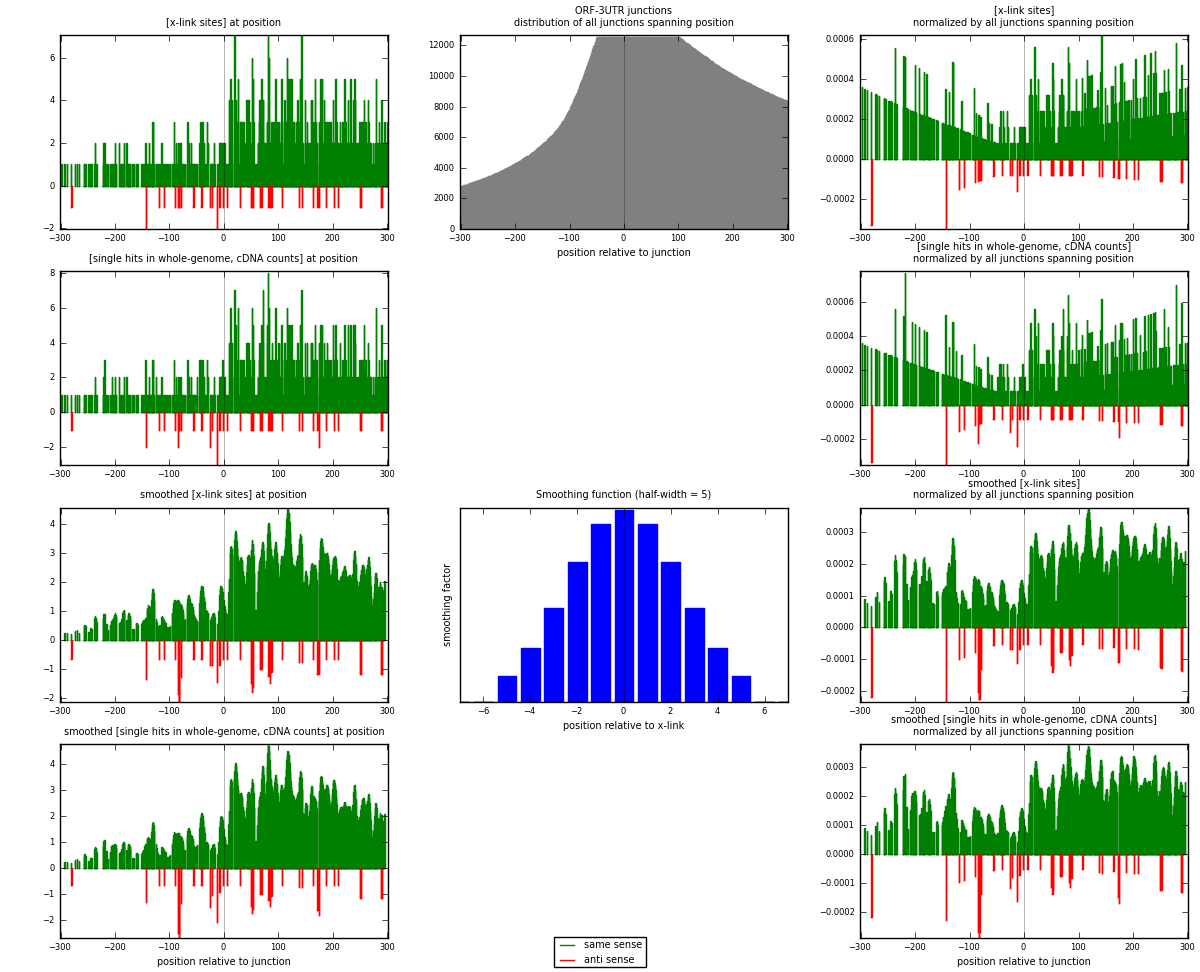 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 813 | 0.65% | 4.22% | 95.20% | 583 | 1.31129 |
| anti | 41 | 0.03% | 0.21% | 4.80% | 37 | 0.066129 |
| both | 854 | 0.68% | 4.43% | 100.00% | 620 | 1.37742 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 858 | 0.65% | 4.10% | 95.02% | 583 | 1.38387 |
| anti | 45 | 0.03% | 0.22% | 4.98% | 37 | 0.0725806 |
| both | 903 | 0.68% | 4.32% | 100.00% | 620 | 1.45645 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
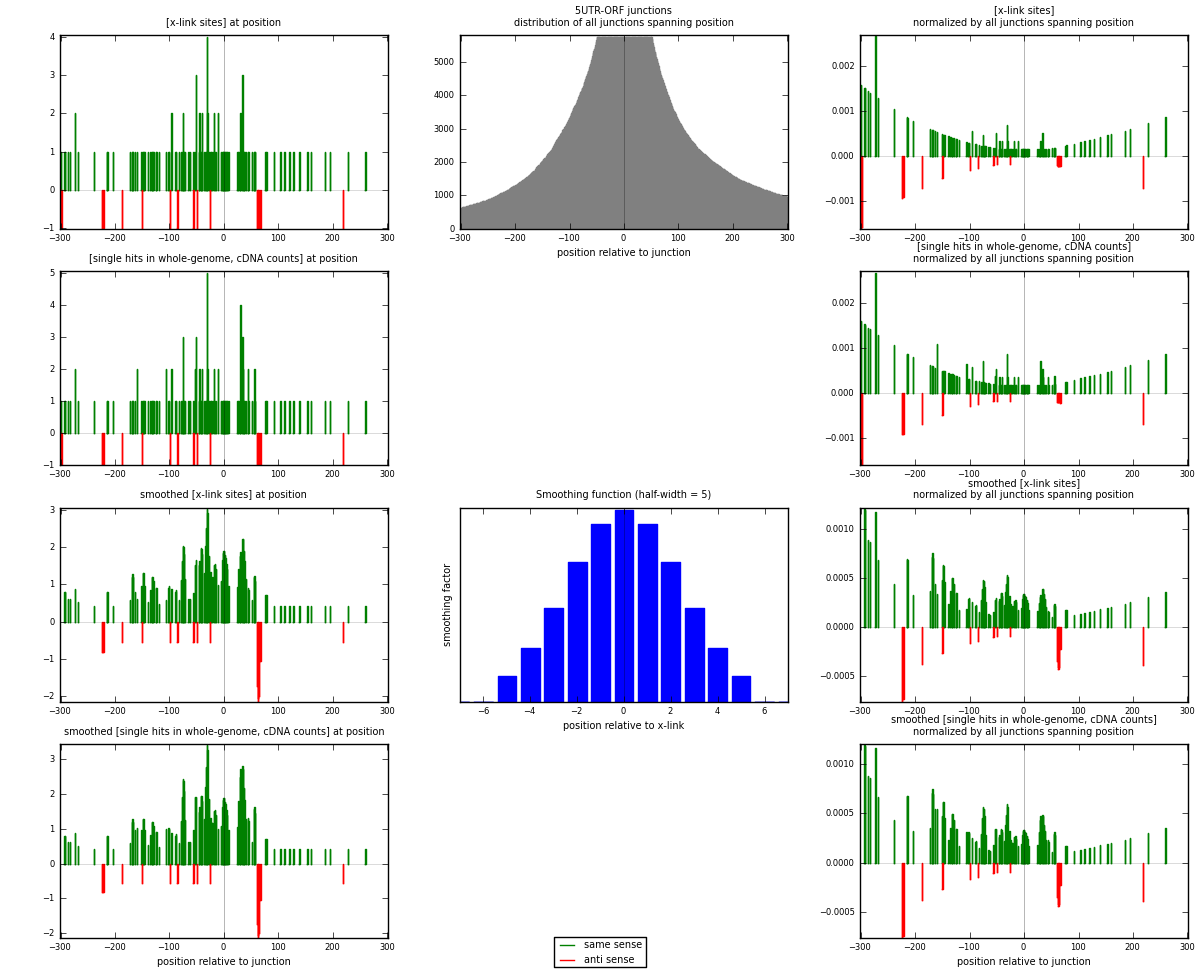 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 120 | 0.10% | 0.62% | 87.59% | 92 | 1.14286 |
| anti | 17 | 0.01% | 0.09% | 12.41% | 13 | 0.161905 |
| both | 137 | 0.11% | 0.71% | 100.00% | 105 | 1.30476 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 130 | 0.10% | 0.62% | 88.44% | 92 | 1.2381 |
| anti | 17 | 0.01% | 0.08% | 11.56% | 13 | 0.161905 |
| both | 147 | 0.11% | 0.70% | 100.00% | 105 | 1.4 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
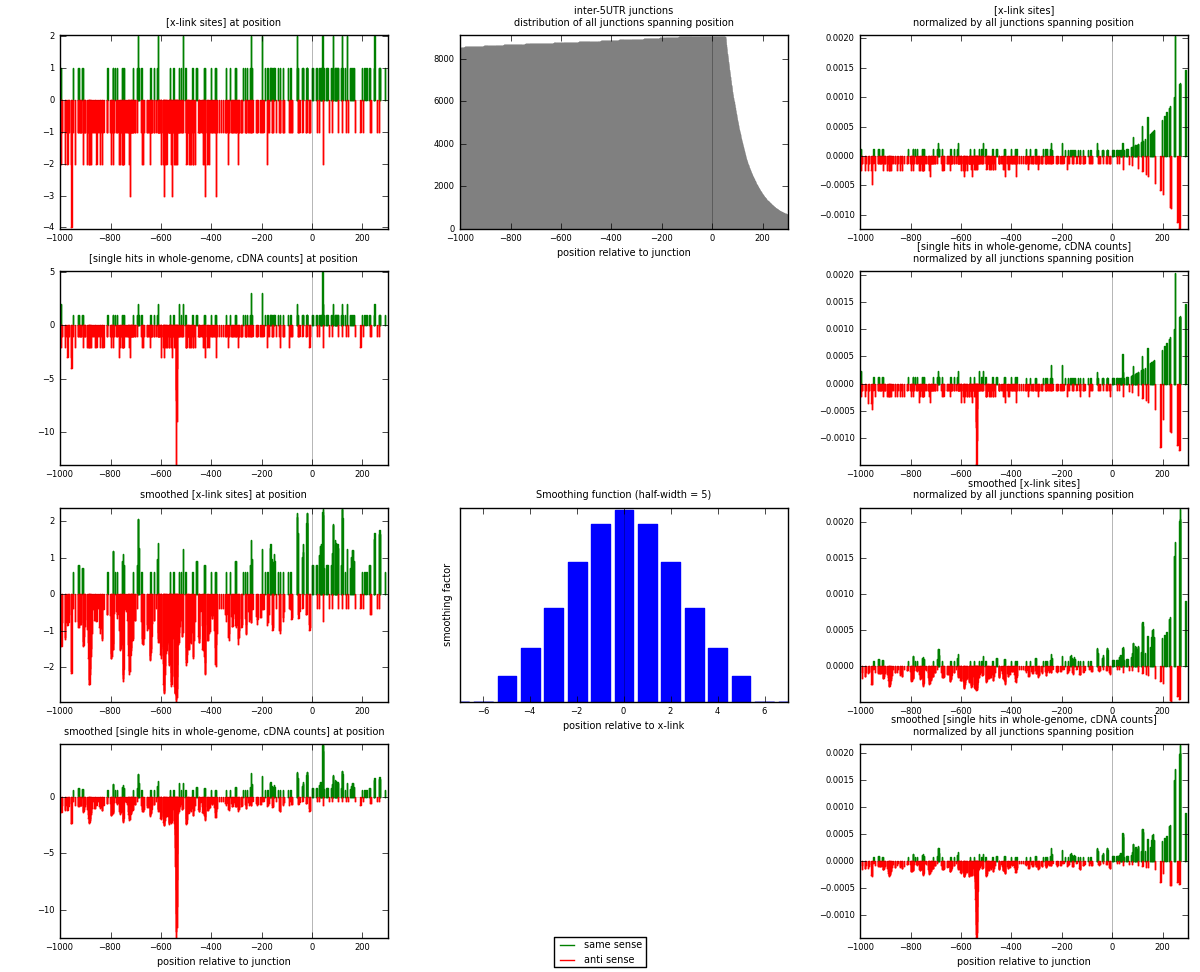 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 129 | 0.10% | 0.67% | 26.27% | 107 | 0.351499 |
| anti | 362 | 0.29% | 1.88% | 73.73% | 262 | 0.986376 |
| both | 491 | 0.39% | 2.55% | 100.00% | 367 | 1.33787 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 137 | 0.10% | 0.65% | 25.05% | 107 | 0.373297 |
| anti | 410 | 0.31% | 1.96% | 74.95% | 262 | 1.11717 |
| both | 547 | 0.41% | 2.61% | 100.00% | 367 | 1.49046 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
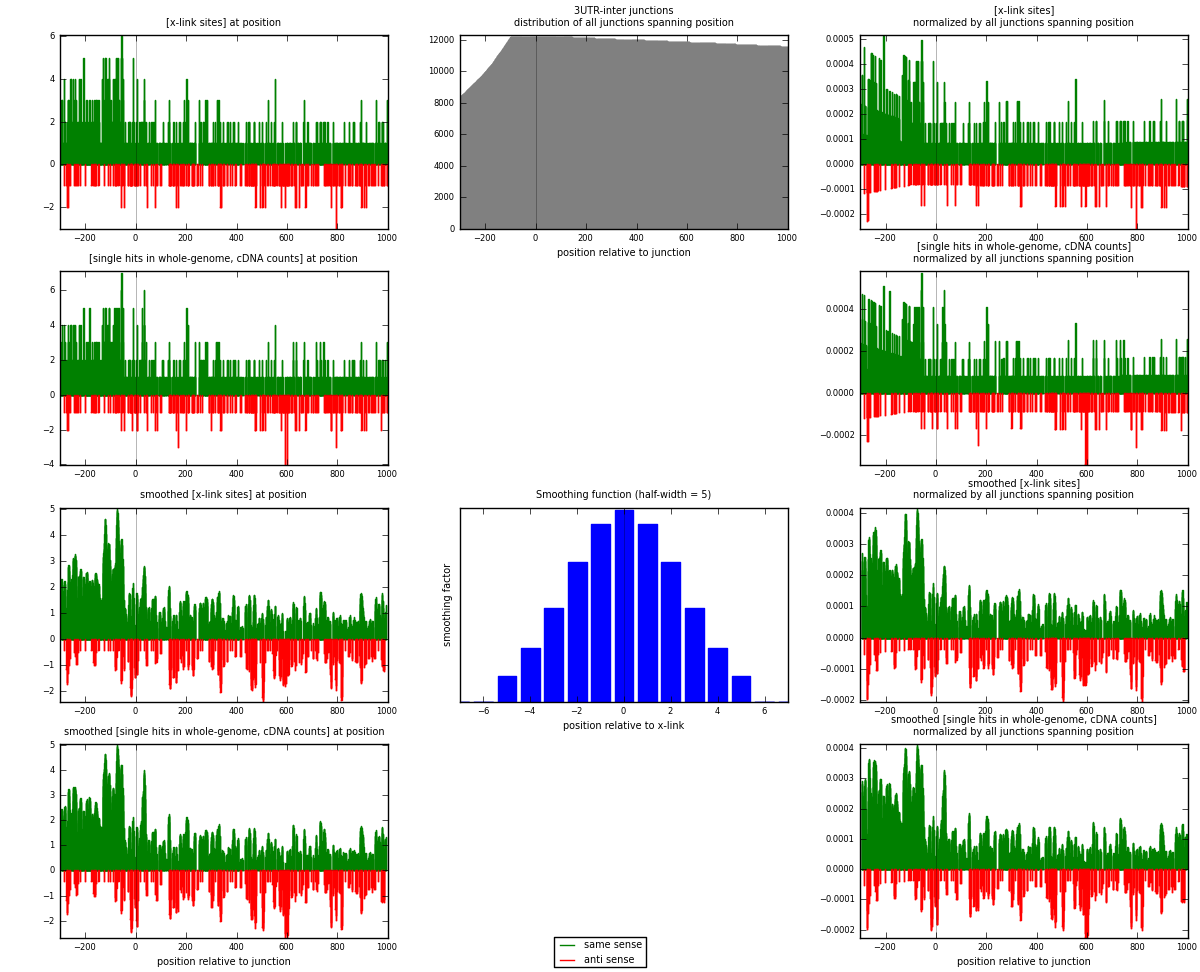 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1011 | 0.81% | 5.24% | 77.41% | 748 | 1.05203 |
| anti | 295 | 0.24% | 1.53% | 22.59% | 225 | 0.306972 |
| both | 1306 | 1.04% | 6.77% | 100.00% | 961 | 1.359 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1073 | 0.81% | 5.13% | 77.58% | 748 | 1.11655 |
| anti | 310 | 0.23% | 1.48% | 22.42% | 225 | 0.322581 |
| both | 1383 | 1.05% | 6.61% | 100.00% | 961 | 1.43913 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
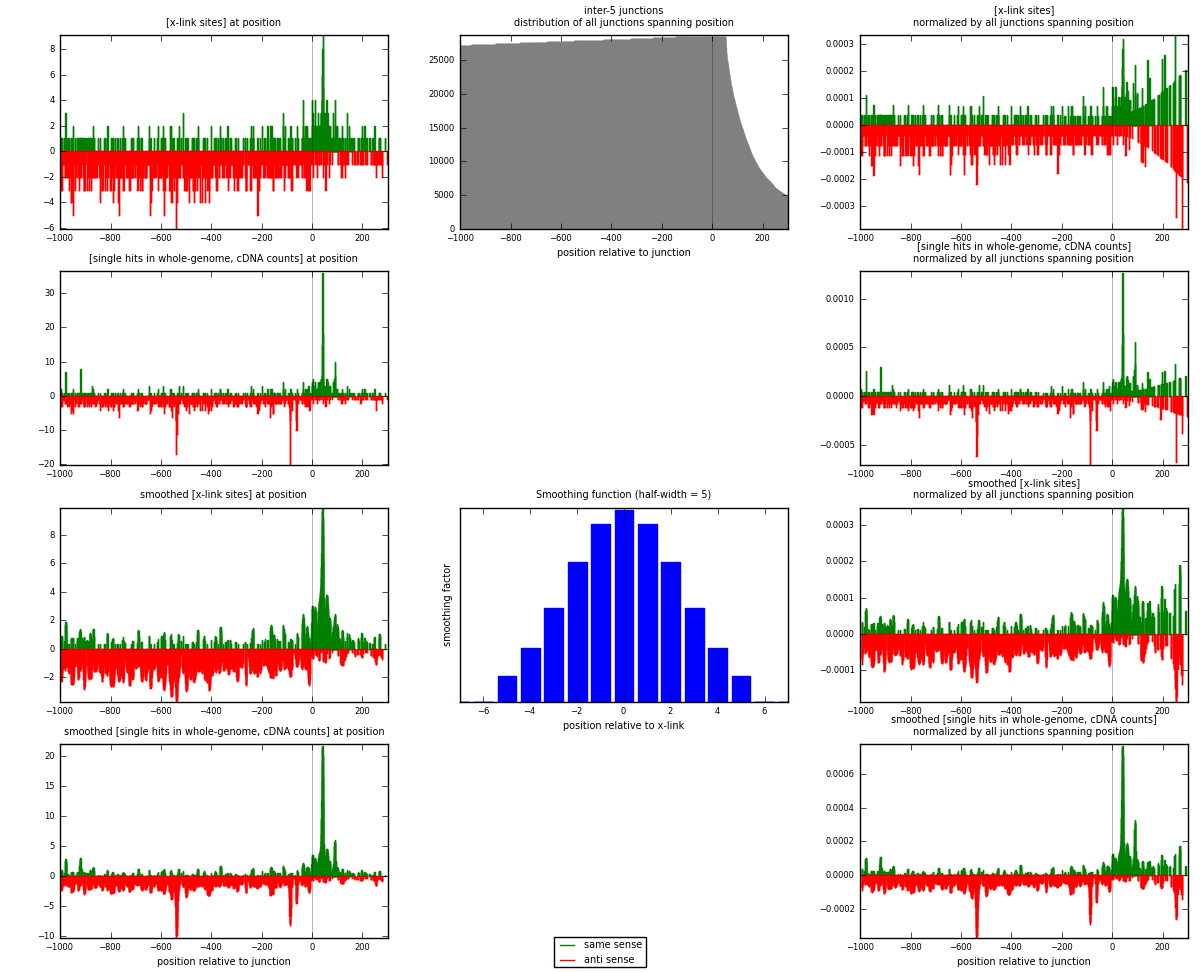 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 446 | 0.36% | 2.31% | 27.96% | 265 | 0.411819 |
| anti | 1149 | 0.92% | 5.96% | 72.04% | 823 | 1.06094 |
| both | 1595 | 1.27% | 8.27% | 100.00% | 1083 | 1.47276 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 563 | 0.43% | 2.69% | 30.29% | 265 | 0.519852 |
| anti | 1296 | 0.98% | 6.20% | 69.71% | 823 | 1.19668 |
| both | 1859 | 1.41% | 8.89% | 100.00% | 1083 | 1.71653 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
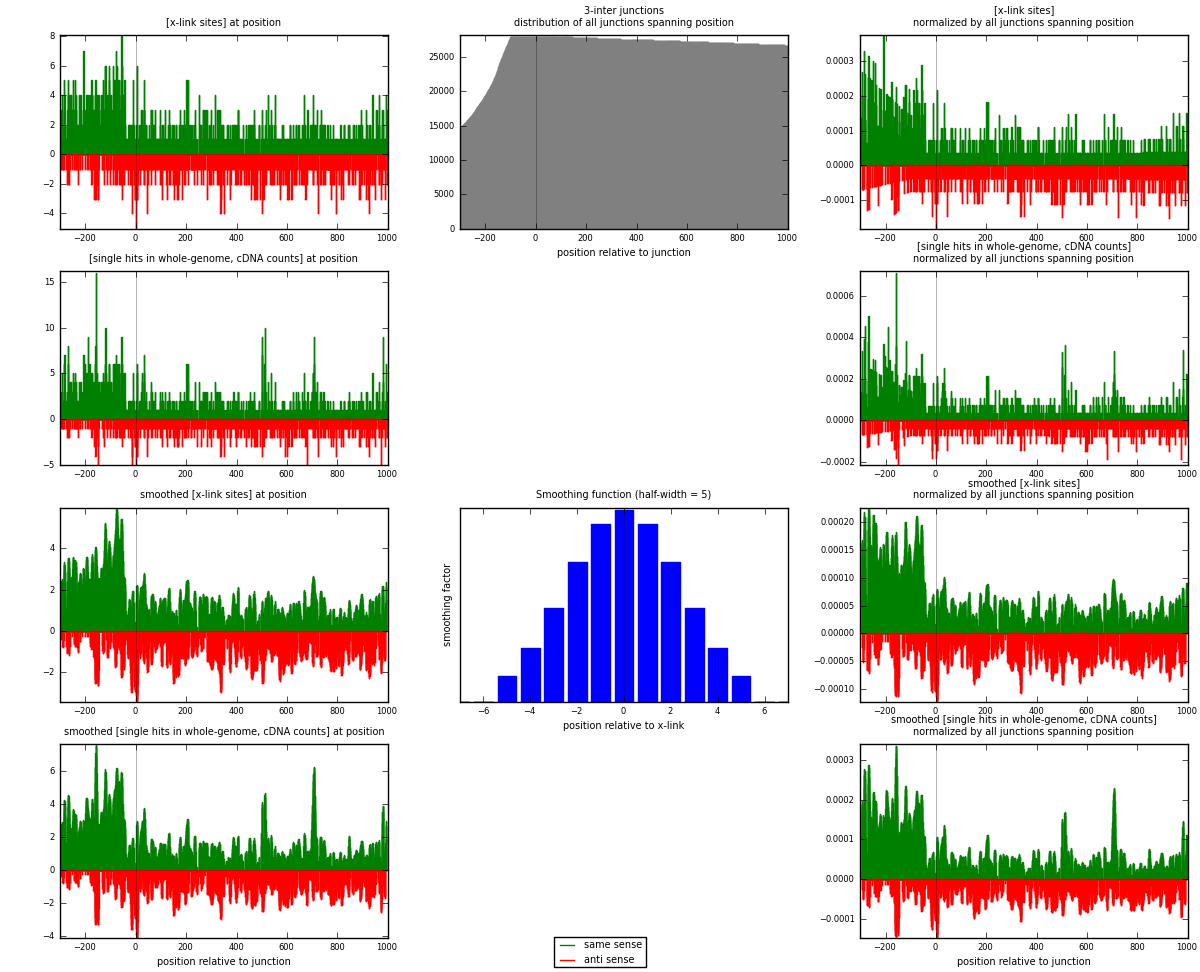 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1477 | 1.18% | 7.66% | 61.64% | 951 | 0.93481 |
| anti | 919 | 0.73% | 4.77% | 38.36% | 648 | 0.581646 |
| both | 2396 | 1.91% | 12.43% | 100.00% | 1580 | 1.51646 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1701 | 1.29% | 8.13% | 63.85% | 951 | 1.07658 |
| anti | 963 | 0.73% | 4.60% | 36.15% | 648 | 0.609494 |
| both | 2664 | 2.02% | 12.73% | 100.00% | 1580 | 1.68608 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
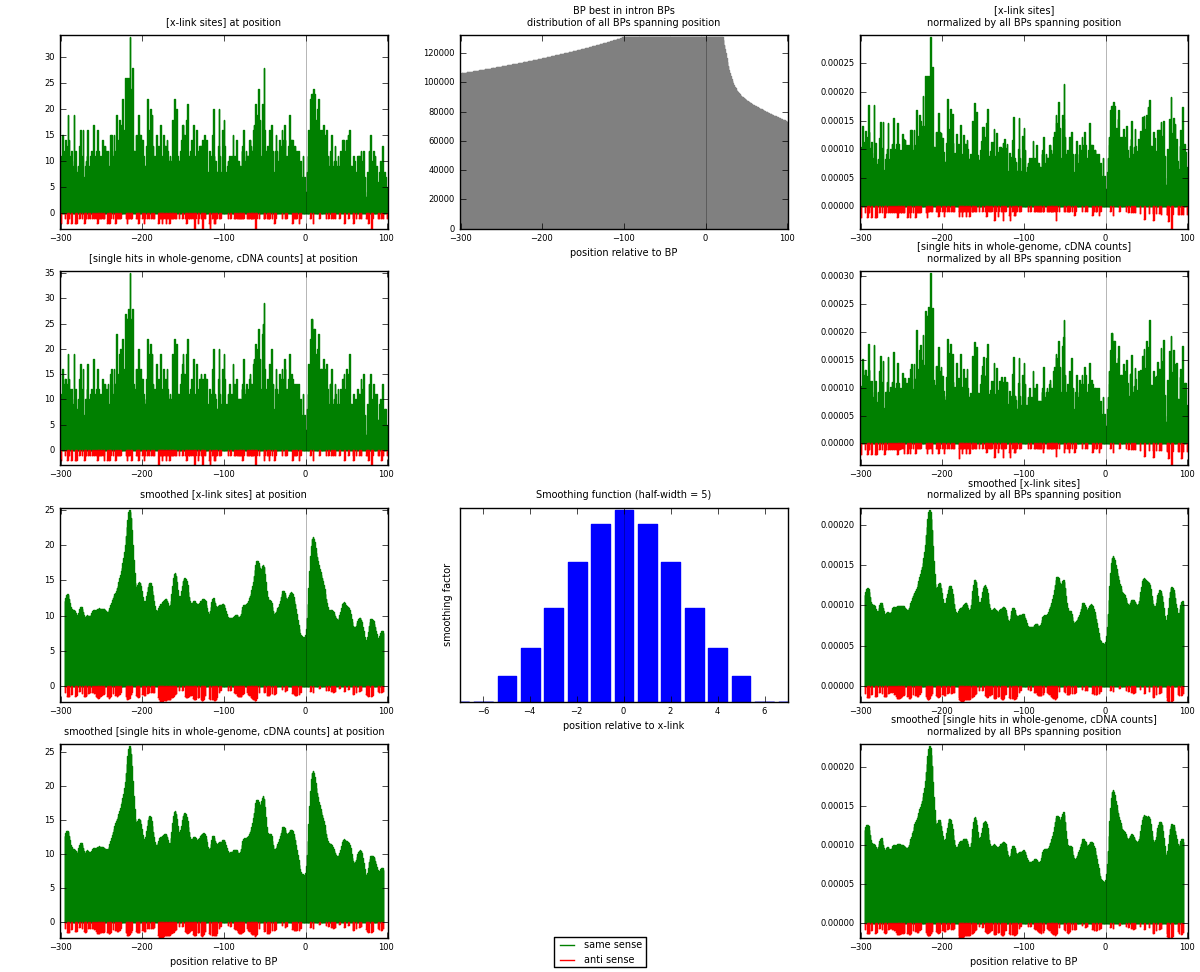 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 4835 | 3.86% | 25.08% | 95.95% | 4038 | 1.14546 |
| anti | 204 | 0.16% | 1.06% | 4.05% | 187 | 0.0483298 |
| both | 5039 | 4.02% | 26.13% | 100.00% | 4221 | 1.19379 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 5051 | 3.82% | 24.14% | 95.99% | 4038 | 1.19664 |
| anti | 211 | 0.16% | 1.01% | 4.01% | 187 | 0.0499882 |
| both | 5262 | 3.98% | 25.15% | 100.00% | 4221 | 1.24662 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.