RNAmap exon-intron
junction is at position zero.upstream is regions of type: ORF, 3UTR, 5UTR
downstream is regions of type: intron
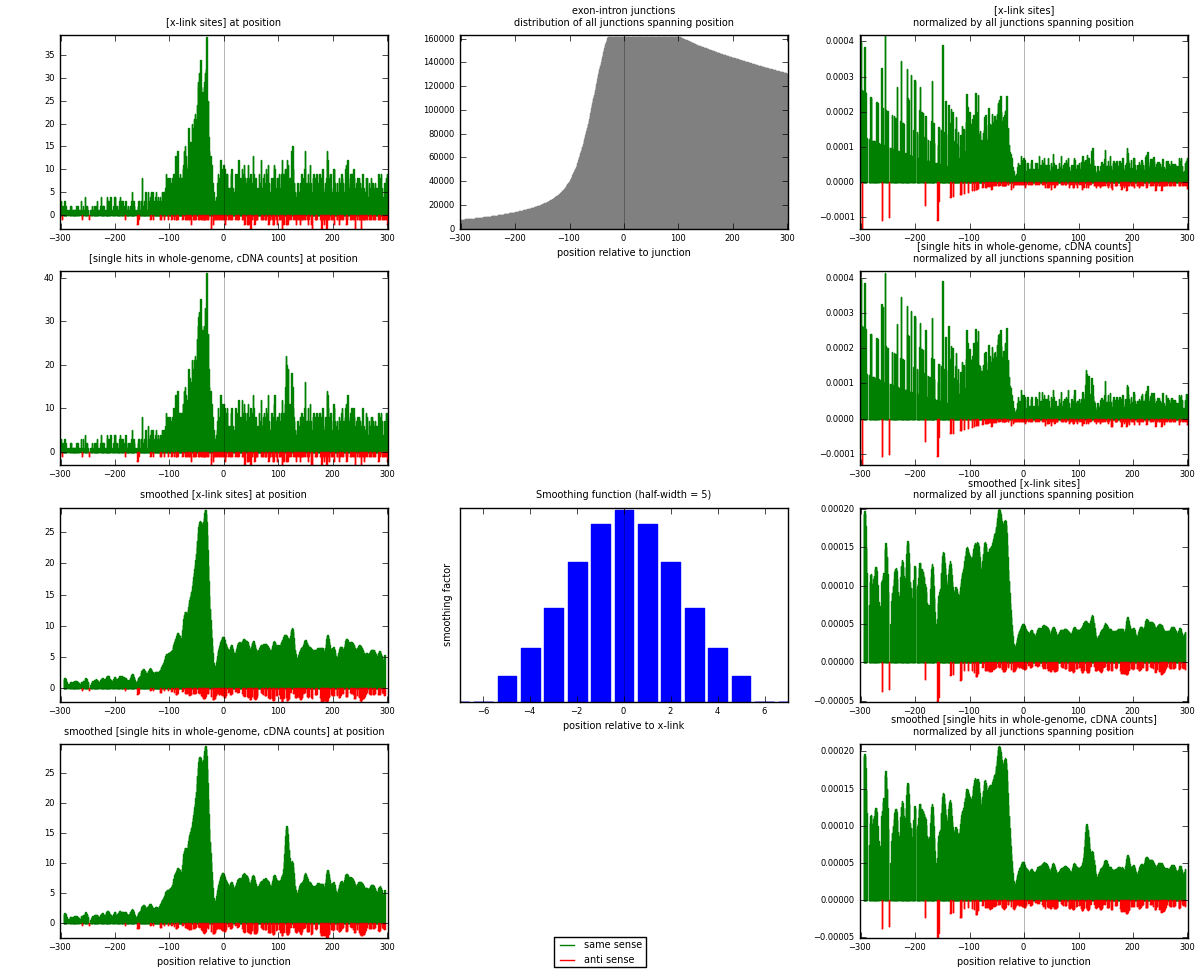 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 3620 | 4.37% | 23.21% | 94.32% | 3348 | 1.01828 |
| anti | 218 | 0.26% | 1.40% | 5.68% | 209 | 0.0613221 |
| both | 3838 | 4.64% | 24.60% | 100.00% | 3555 | 1.07961 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3798 | 4.21% | 19.85% | 94.43% | 3348 | 1.06835 |
| anti | 224 | 0.25% | 1.17% | 5.57% | 209 | 0.0630098 |
| both | 4022 | 4.46% | 21.02% | 100.00% | 3555 | 1.13136 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap intron-exon
junction is at position zero.upstream is regions of type: intron
downstream is regions of type: ORF, 3UTR, 5UTR
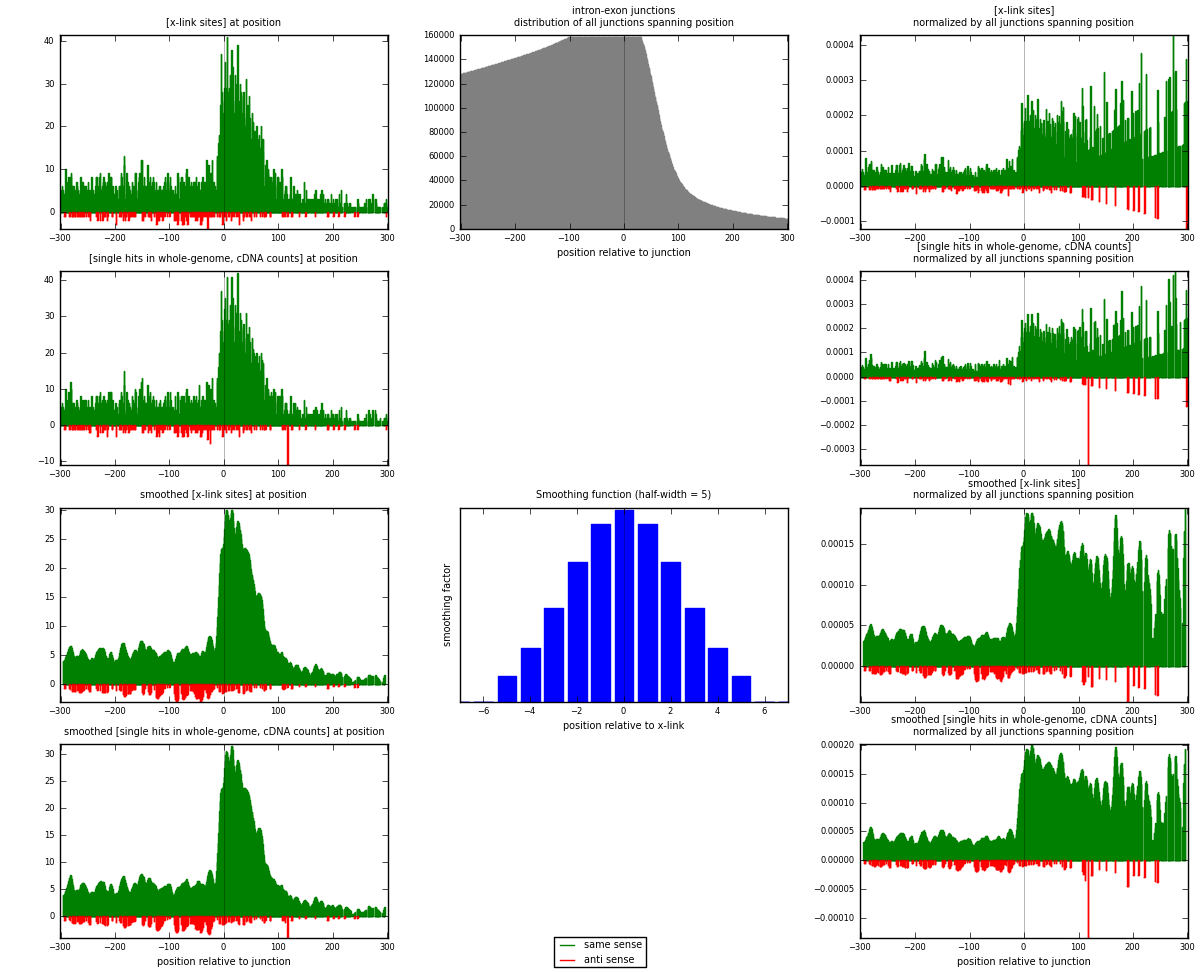 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 4038 | 4.88% | 25.88% | 95.03% | 3763 | 1.02073 |
| anti | 211 | 0.25% | 1.35% | 4.97% | 199 | 0.0533367 |
| both | 4249 | 5.13% | 27.24% | 100.00% | 3956 | 1.07406 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 4154 | 4.61% | 21.71% | 94.73% | 3763 | 1.05005 |
| anti | 231 | 0.26% | 1.21% | 5.27% | 199 | 0.0583923 |
| both | 4385 | 4.86% | 22.92% | 100.00% | 3956 | 1.10844 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap 5-ncRNA
junction is at position zero.upstream is regions of type: intron, inter, telo
downstream is regions of type: ncRNA
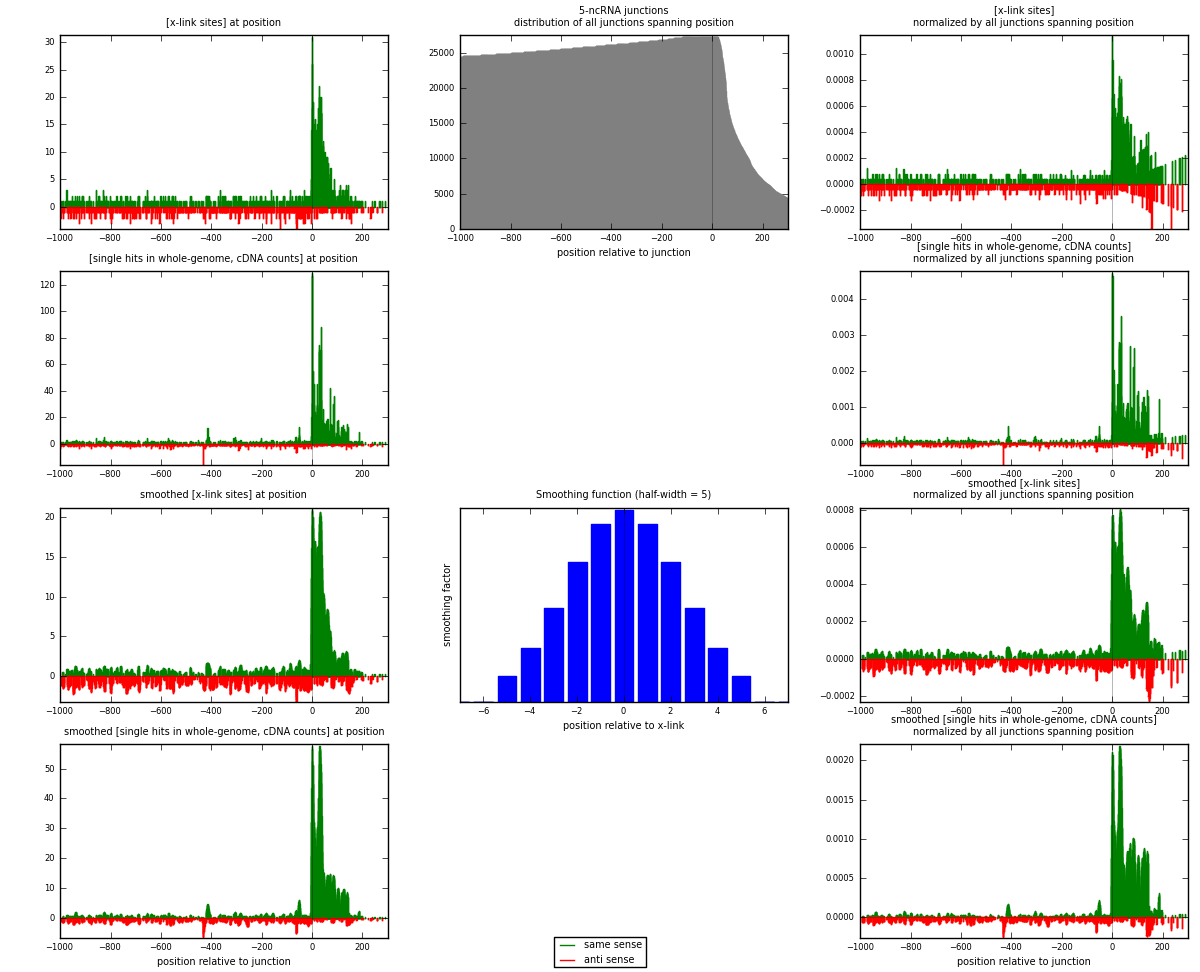 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1513 | 1.83% | 9.70% | 72.36% | 601 | 1.42333 |
| anti | 578 | 0.70% | 3.71% | 27.64% | 472 | 0.543744 |
| both | 2091 | 2.53% | 13.40% | 100.00% | 1063 | 1.96707 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 3028 | 3.36% | 15.82% | 82.78% | 601 | 2.84854 |
| anti | 630 | 0.70% | 3.29% | 17.22% | 472 | 0.592662 |
| both | 3658 | 4.06% | 19.12% | 100.00% | 1063 | 3.4412 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
RNAmap ncRNA-3
junction is at position zero.upstream is regions of type: ncRNA
downstream is regions of type: intron, inter, telo
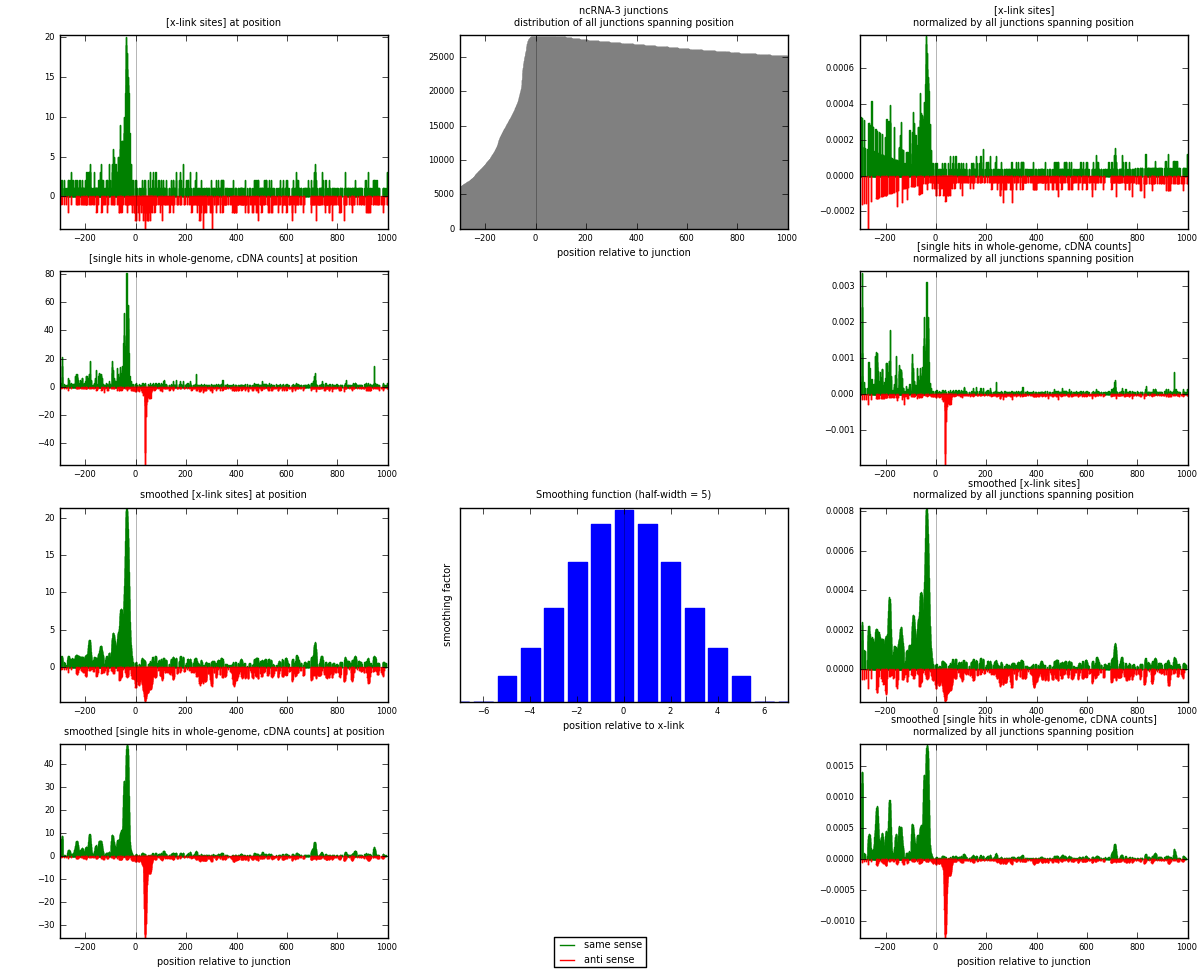 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1192 | 1.44% | 7.64% | 68.51% | 485 | 1.37963 |
| anti | 548 | 0.66% | 3.51% | 31.49% | 384 | 0.634259 |
| both | 1740 | 2.10% | 11.15% | 100.00% | 864 | 2.01389 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2167 | 2.40% | 11.32% | 73.28% | 485 | 2.5081 |
| anti | 790 | 0.88% | 4.13% | 26.72% | 384 | 0.914352 |
| both | 2957 | 3.28% | 15.45% | 100.00% | 864 | 3.42245 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
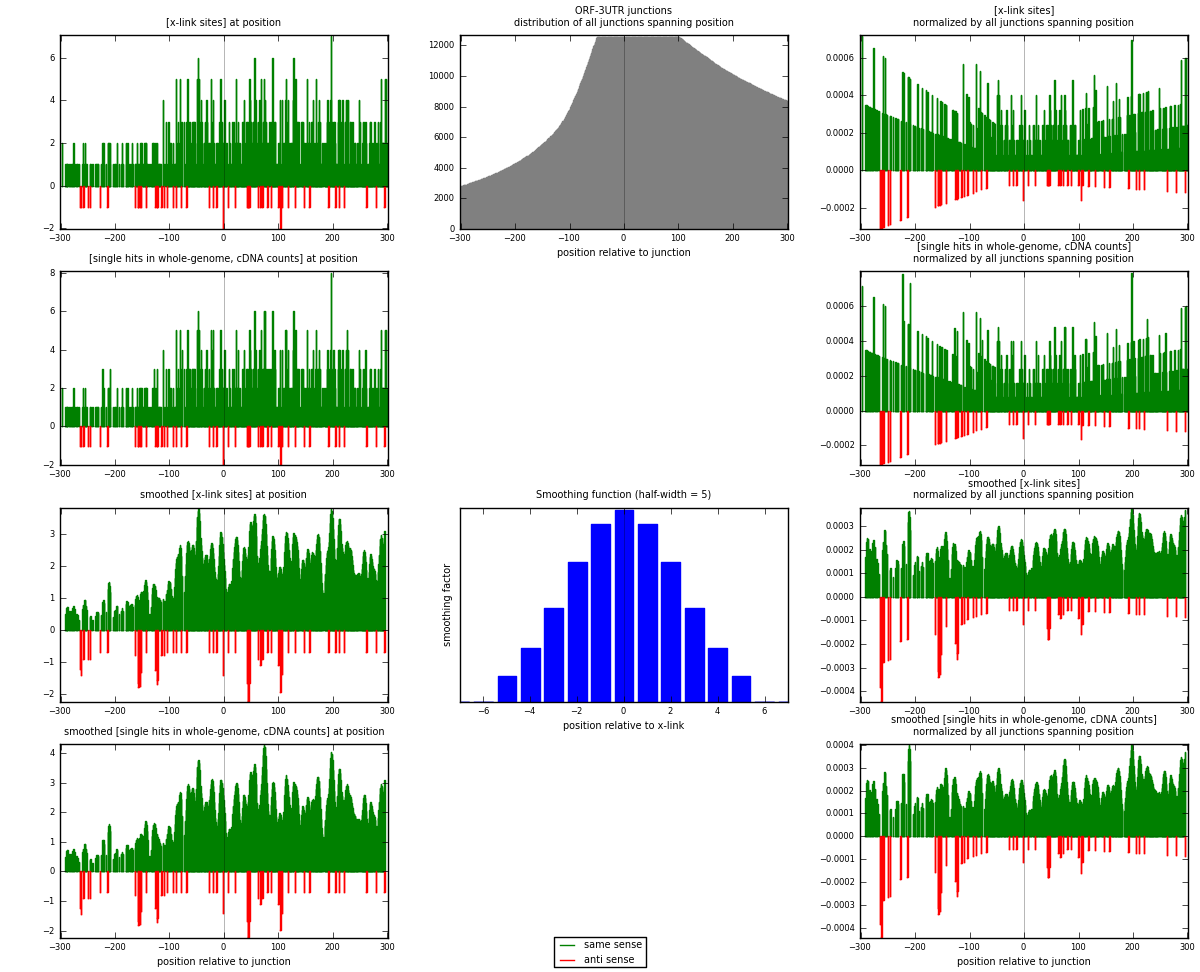
RNAmap ORF-3UTR
junction is at position zero.upstream is regions of type: ORF
downstream is regions of type: 3UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 914 | 1.10% | 5.86% | 94.42% | 742 | 1.17029 |
| anti | 54 | 0.07% | 0.35% | 5.58% | 42 | 0.0691421 |
| both | 968 | 1.17% | 6.21% | 100.00% | 781 | 1.23944 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 943 | 1.05% | 4.93% | 94.58% | 742 | 1.20743 |
| anti | 54 | 0.06% | 0.28% | 5.42% | 42 | 0.0691421 |
| both | 997 | 1.11% | 5.21% | 100.00% | 781 | 1.27657 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
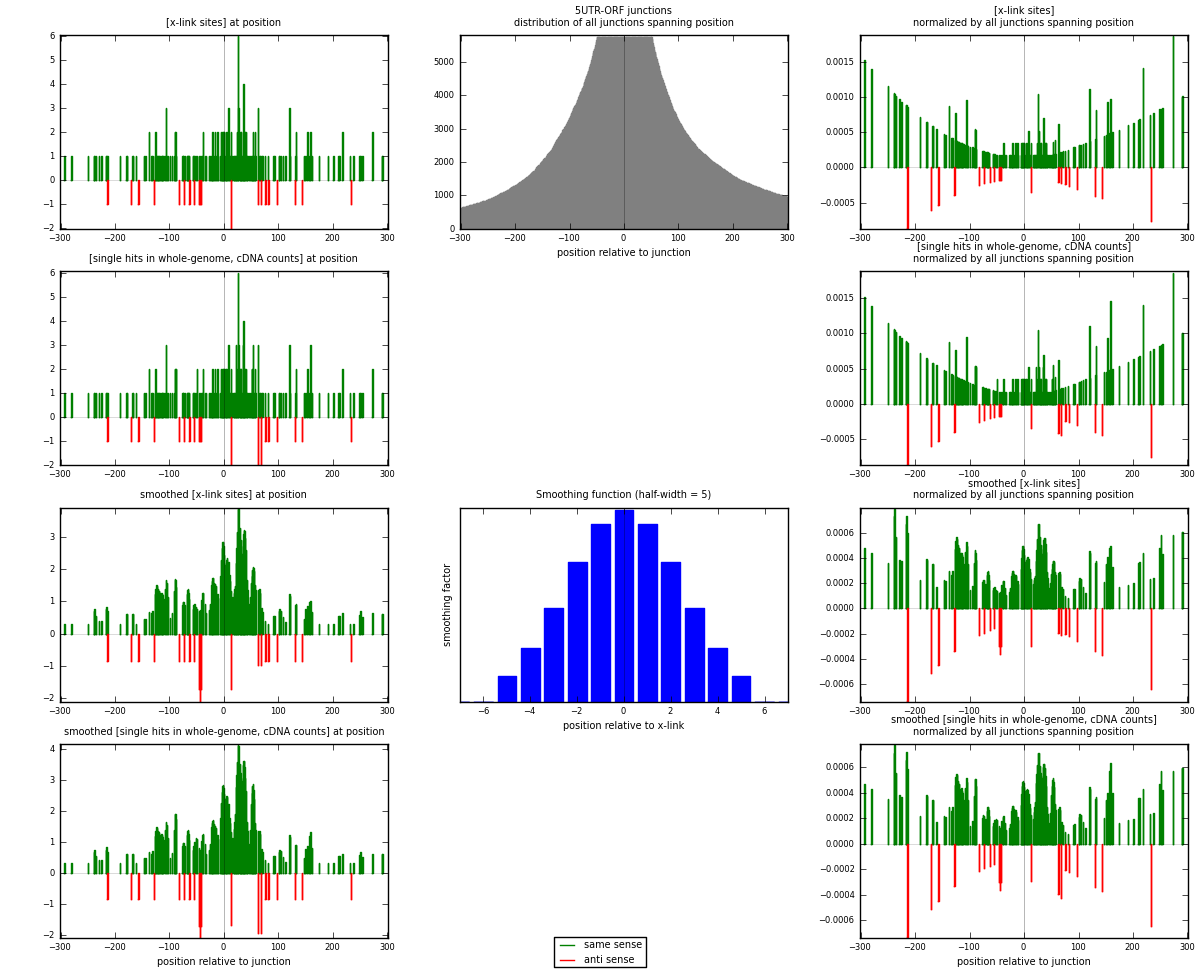
RNAmap 5UTR-ORF
junction is at position zero.upstream is regions of type: 5UTR
downstream is regions of type: ORF
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 208 | 0.25% | 1.33% | 90.83% | 168 | 1.13661 |
| anti | 21 | 0.03% | 0.13% | 9.17% | 15 | 0.114754 |
| both | 229 | 0.28% | 1.47% | 100.00% | 183 | 1.25137 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 220 | 0.24% | 1.15% | 90.53% | 168 | 1.20219 |
| anti | 23 | 0.03% | 0.12% | 9.47% | 15 | 0.125683 |
| both | 243 | 0.27% | 1.27% | 100.00% | 183 | 1.32787 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
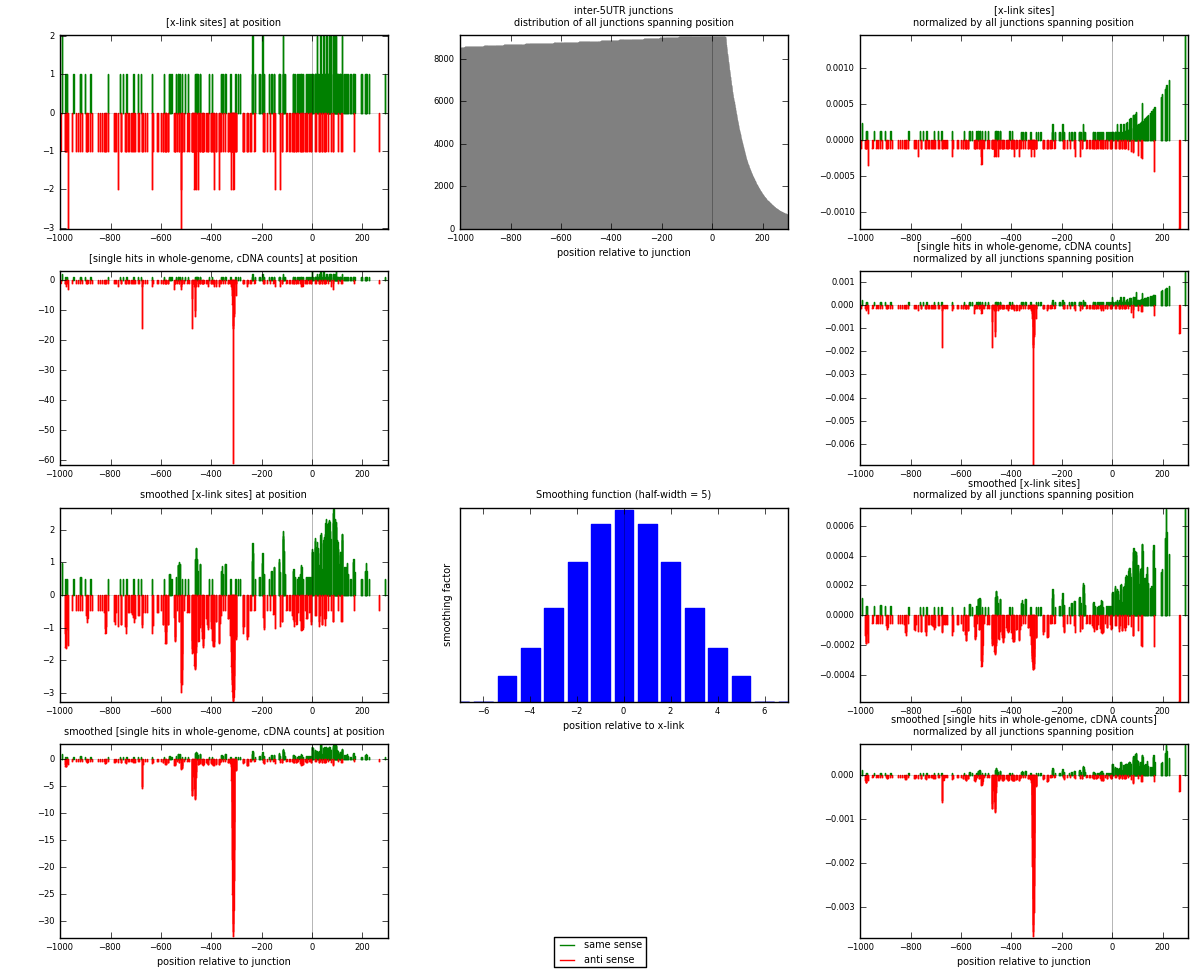
RNAmap inter-5UTR
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: 5UTR
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 165 | 0.20% | 1.06% | 44.00% | 148 | 0.559322 |
| anti | 210 | 0.25% | 1.35% | 56.00% | 151 | 0.711864 |
| both | 375 | 0.45% | 2.40% | 100.00% | 295 | 1.27119 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 172 | 0.19% | 0.90% | 29.40% | 148 | 0.583051 |
| anti | 413 | 0.46% | 2.16% | 70.60% | 151 | 1.4 |
| both | 585 | 0.65% | 3.06% | 100.00% | 295 | 1.98305 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
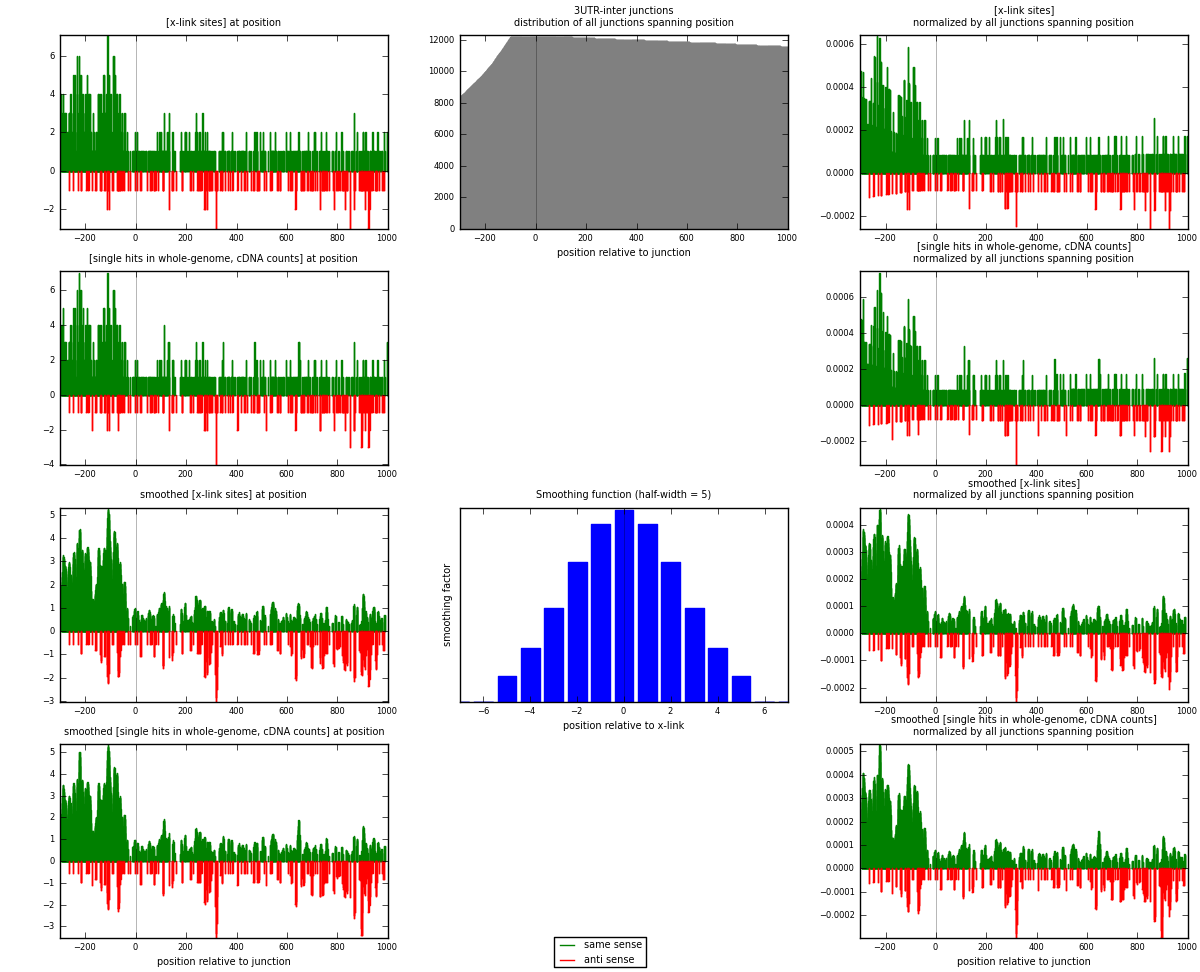
RNAmap 3UTR-inter
junction is at position zero.upstream is regions of type: 3UTR
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 891 | 1.08% | 5.71% | 81.89% | 749 | 0.992205 |
| anti | 197 | 0.24% | 1.26% | 18.11% | 156 | 0.219376 |
| both | 1088 | 1.31% | 6.97% | 100.00% | 898 | 1.21158 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 913 | 1.01% | 4.77% | 81.52% | 749 | 1.0167 |
| anti | 207 | 0.23% | 1.08% | 18.48% | 156 | 0.230512 |
| both | 1120 | 1.24% | 5.85% | 100.00% | 898 | 1.24722 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
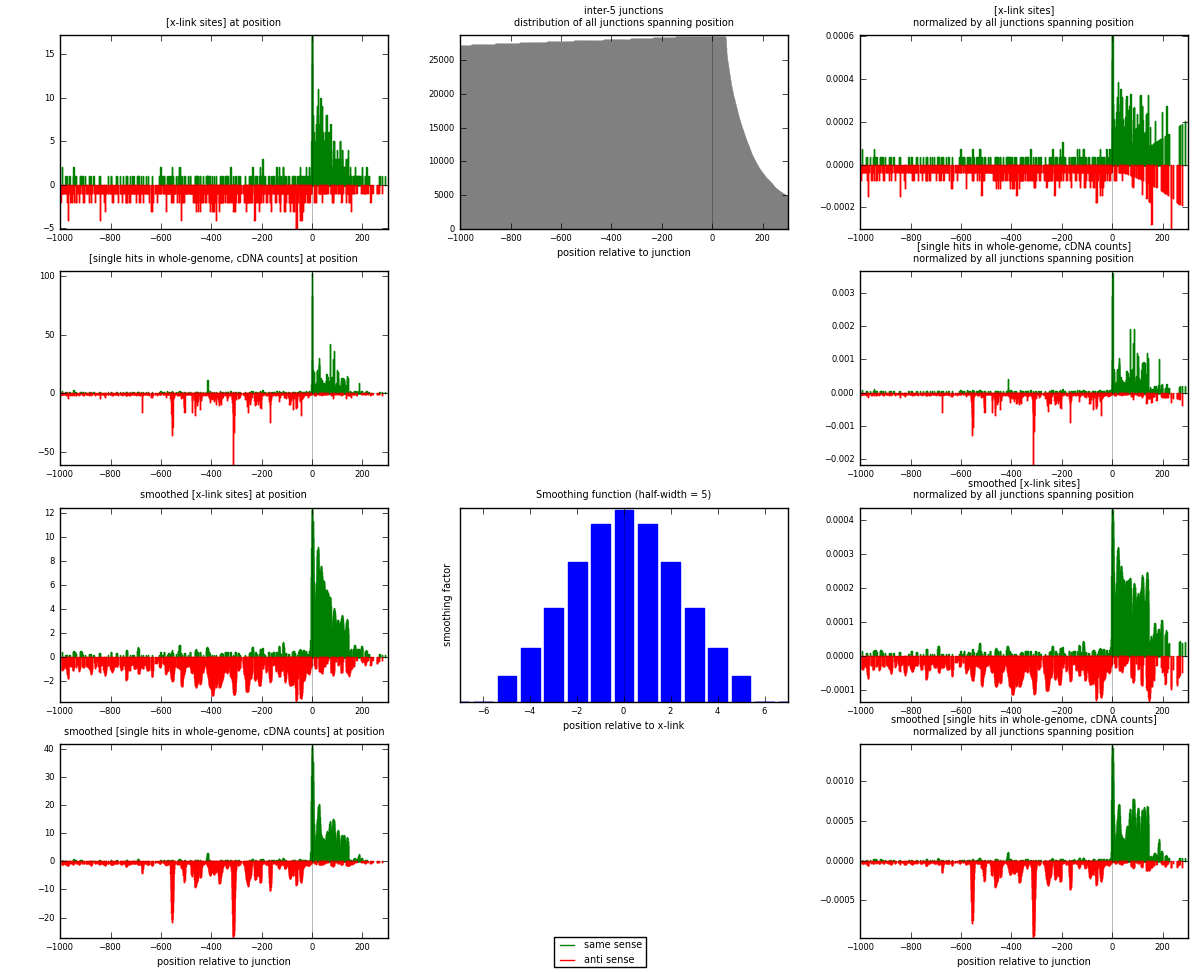
RNAmap inter-5
junction is at position zero.upstream is regions of type: inter, telo
downstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 781 | 0.94% | 5.01% | 46.74% | 413 | 0.829087 |
| anti | 890 | 1.08% | 5.71% | 53.26% | 538 | 0.944798 |
| both | 1671 | 2.02% | 10.71% | 100.00% | 942 | 1.77389 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 1637 | 1.82% | 8.56% | 47.34% | 413 | 1.73779 |
| anti | 1821 | 2.02% | 9.52% | 52.66% | 538 | 1.93312 |
| both | 3458 | 3.83% | 18.07% | 100.00% | 942 | 3.67091 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
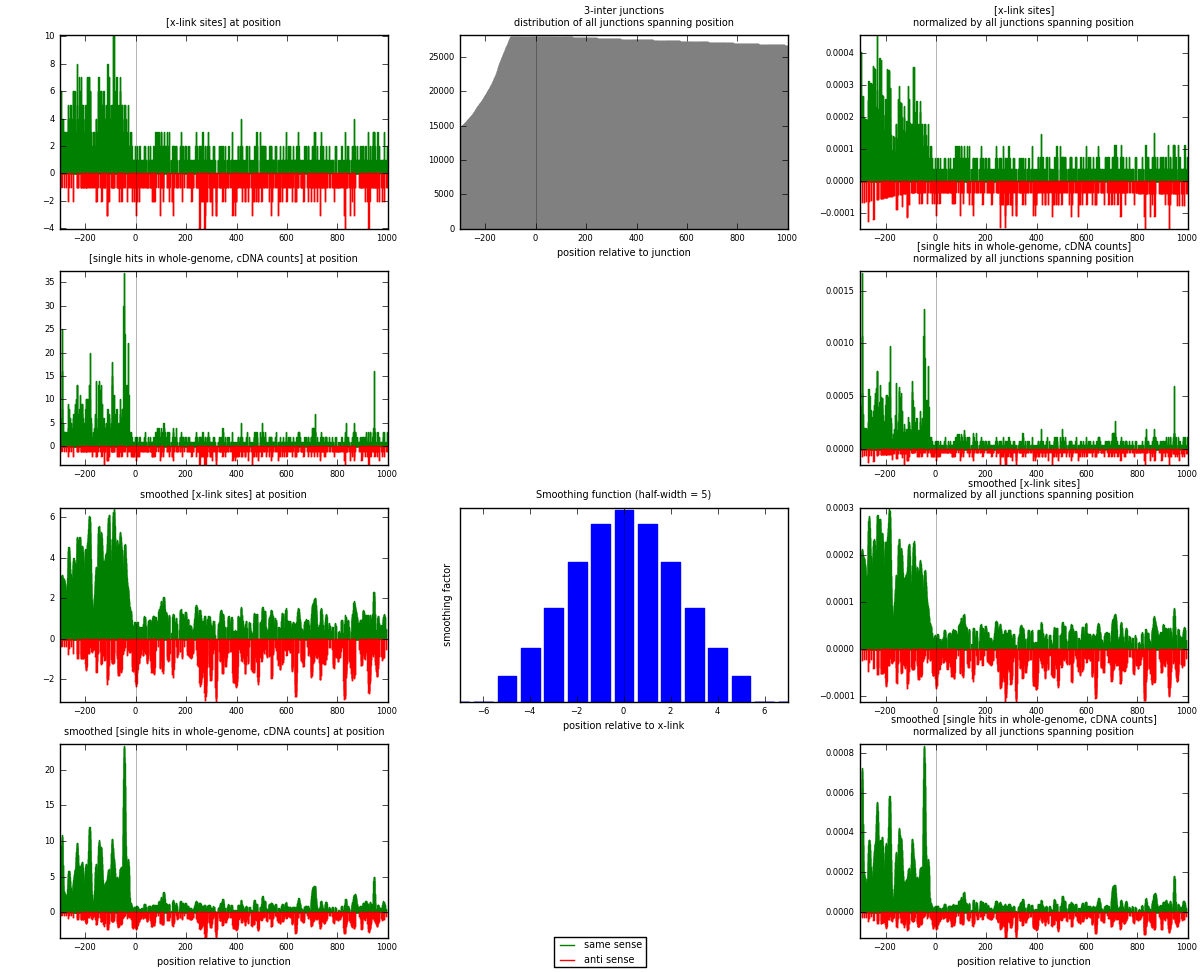
RNAmap 3-inter
junction is at position zero.upstream is regions of type: ORF, intron, 3UTR, 5UTR, ncRNA
downstream is regions of type: inter, telo
 [eps]
[eps]x-links and junctions distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [junctions] in map | [x-link sites] per unique [junction] |
|---|---|---|---|---|---|---|
| same | 1554 | 1.88% | 9.96% | 73.20% | 954 | 1.13846 |
| anti | 569 | 0.69% | 3.65% | 26.80% | 419 | 0.41685 |
| both | 2123 | 2.56% | 13.61% | 100.00% | 1365 | 1.55531 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [junctions] in map | [single hits in whole-genome, cDNA counts] per unique [junction] |
| same | 2201 | 2.44% | 11.50% | 78.50% | 954 | 1.61245 |
| anti | 603 | 0.67% | 3.15% | 21.50% | 419 | 0.441758 |
| both | 2804 | 3.11% | 14.65% | 100.00% | 1365 | 2.05421 |
Top 10 junctions with largest x-link sites sum (both strands)
|
Top 10 junctions with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.
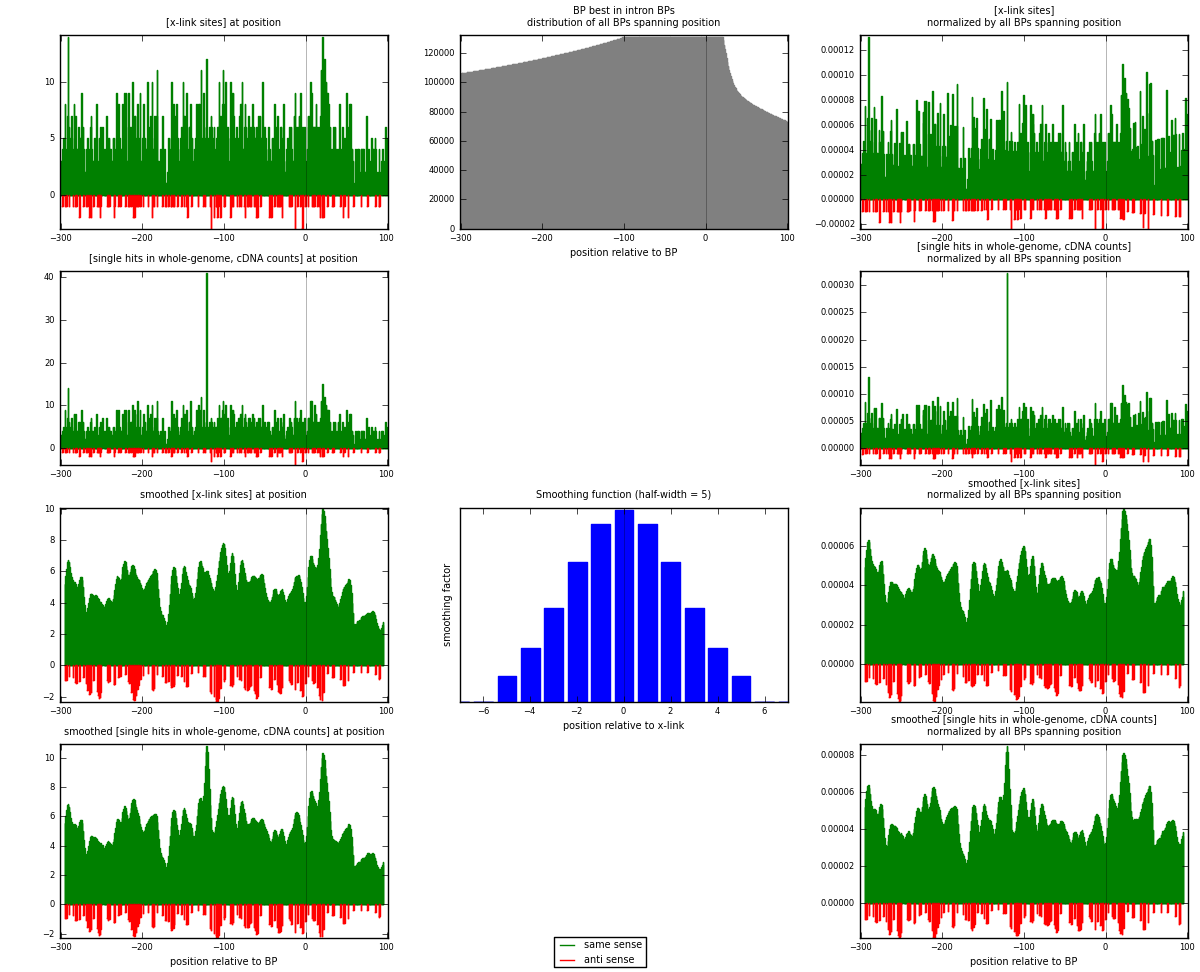
RNAmap "BP best in intron"
BP is at position zero.upstream is usptream of branch point
downstream is downstream of branch point
 [eps]
[eps]x-links and BPs distributions
| strand | [x-link sites] used in map | [x-link sites] % of total | [x-link sites] % of used in any map | [x-link sites] % of used in map | unique [BPs] in map | [x-link sites] per unique [BP] |
|---|---|---|---|---|---|---|
| same | 2013 | 2.43% | 12.90% | 92.85% | 1748 | 1.06003 |
| anti | 155 | 0.19% | 0.99% | 7.15% | 151 | 0.0816219 |
| both | 2168 | 2.62% | 13.90% | 100.00% | 1899 | 1.14165 |
| strand | [single hits in whole-genome, cDNA counts] used in map | [single hits in whole-genome, cDNA counts] % of total | [single hits in whole-genome, cDNA counts] % of used in any map | [single hits in whole-genome, cDNA counts] % of used in map | unique [BPs] in map | [single hits in whole-genome, cDNA counts] per unique [BP] |
| same | 2119 | 2.35% | 11.07% | 93.06% | 1748 | 1.11585 |
| anti | 158 | 0.18% | 0.83% | 6.94% | 151 | 0.0832017 |
| both | 2277 | 2.52% | 11.90% | 100.00% | 1899 | 1.19905 |
Top 10 BPs with largest x-link sites sum (both strands)
|
Top 10 BPs with largest [single hits in whole-genome, cDNA counts] sum (both strands)
|
Download full list of x-links used for RNAmap in tab-delimited format.